-- stem_cell_rna
|__stem_cell_rna.Rproj
|__raw_ data/
|__2019-03-21_donor_1.csv
|__2019-03-21_donor_2.csv
|__2019-03-21_donor_3.csv
|__README.md
|__R/
|__01_data_processing.R
|__02_exploratory.R
|__functions/
|__theme_volcano.R
|__normalise.RWorkshop
Supporting Information 1 Organising Reproducible Data Analyses
3 December, 2025
Introduction
Session overview
In this workshop we will discuss why reproducibility matters and how to organise your work to make it reproducible. We will cover:
- What is reproducibility
- How to achieve reproducibility
- Rationale for scripting
- Project-oriented workflow
- Code formatting and style
- Coding algorithmically
- Naming things
- And some handy workflow tips
Slide navigation
Reproducibility
What is reproducibility?
Definitions
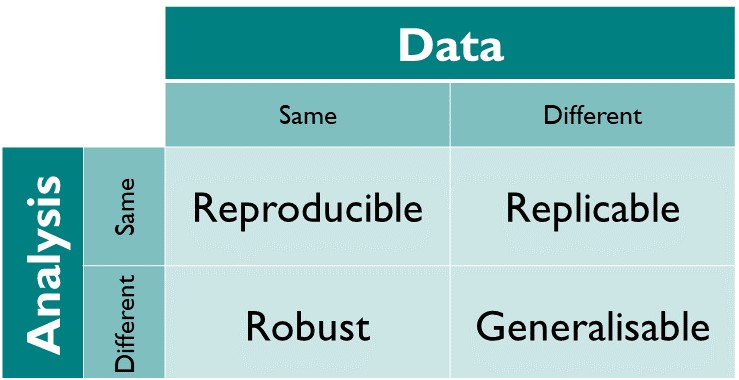
Reproducible: Same data + same analysis = identical results. “… obtaining consistent results using the same input data; computational steps, methods, and code; and conditions of analysis. This definition is synonymous with”computational reproducibility” (National Academies of Sciences et al. 2019). This is what we are concentrating on in the Supporting Information.
Definitions
Replicable: Different data + same analysis = qualitatively similar results. The work is not dependent on the specificities of the data.
Definitions
Robust: Same data + different analysis = qualitatively similar or identical results. The work is not dependent on the specificities of the analysis.
Definitions
Generalisable: Different data + different analysis = qualitatively similar results and same conclusions.
Why does it matter?
Many high profile cases of work which did not reproduce e.g. Anil Potti unravelled by Baggerly and Coombes (2009)
Five selfish reasons to work reproducibly (Markowetz 2015). Alternatively, see the very entertaining talk
Will become standard in Science and publishing e.g OECD Global Science Forum Building digital workforce capacity and skills for data-intensive science (OECD Global Science Forum 2020)
How to achieve reproducibility
Reproducibility is a continuum. Some is better than none!
Script everything
Organisation: Project-oriented workflows with file and folder structure, naming things
Code: follow a consistent style, organise into sections and scripts (be modular), Code algorithmically
Documentation: Readme files, code comments, metadata,
More advanced: version, control, continuous integration and testing (not required for Supporting Information)
Scripting
Rationale for scripting
Science is the generation of ideas, designing work to test them and reporting the results.
We ensure laboratory and field work is replicable, robust and generalisable by planning and recording in lab books and using standard protocols. Repeating results is still hard.
Workflows for computational projects, and the data analysis and reporting of other work can, and should, be 100% reproducible!
Scripting is the way to achieve this.
Organisation of Supporting Information
Project-oriented workflow
use folders to organise your work
you are aiming for structured, systematic and repeatable.
inputs and outputs should be clearly identifiable from structure and/or naming
Example: SI itself is an RSP
Example: SI includes an RSP
-- stem_cell_rna
|__data_processing/
|__01_data_processing.py
|__02_exploratory.py
|__raw_data/
|__2019-03-21_donor_1.csv
|__2019-03-21_donor_2.csv
|__2019-03-21_donor_3.csv
|__README.md
|__statistical_analysis
|__statistical_analysis.Rproj
|__processed_data/
|__R/
|__01_DGE.R
|__02_visualisation.R
|__functions/
|__theme_volcano.R
|__normalise.RRStudio Projects revisited
RStudio Projects
- RStudio Projects make it easy to manage working directories and paths because they set the working directory to the RStudio Projects directory automatically.
RStudio Projects
RStudio Projects
the .RProj file is directly under the project folder1. Its presence is what makes the folder an RStudio Project
RStudio Projects
When you open an RStudio Project, the working directory is set to the Project directory (i.e., the location of the
.Rprojfile).When you use an RStudio Project you do not need to use
setwd()When someone, including future you, opens the project on another machine, all the paths just work.
RStudio Projects

Jenny Bryan
In the words of Jenny Bryan:
“If the first line of your R script is setwd(”C:/Users/jenny/path/that/only/I/have”) I will come into your office and SET YOUR COMPUTER ON FIRE”
Creating an RStudio Project
There are two menus options:
Top left, File menu
Top Right, drop-down indicated by the
.RProjicon
They both do the same thing.
Creating an RStudio Project
Then Choose: New Project | New Directory | New Project
Make sure you “Browse” to the folder you want to create the project.
❔ Is your working directory a good place to create a Project folder?
Creating an RStudio Project
When you create a new RStudio Project
- A folder called
bananas/is created - RStudio starts a new session in
bananas/i.e., your working directory is nowbananas/ - A file called
bananas.Rprojis created - the
.Rprojfile is what makes the directory an RStudio Project
Opening and closing
You can close an RStudio Project with ONE of:
- File | Close Project
- Using the drop-down option on the far right of the tool bar where you see the Project name
Opening and closing
You can open an RStudio Project with ONE of:
- File | Open Project or File | Recent Projects
- Using the drop-down option on the far right of the tool bar where you see the Project name
- Double-clicking an .Rproj file from your file explorer/finder
When you open project, a new R session starts.
Code formatting and style
Code formatting and style
“Good coding style is like correct punctuation: you can manage without it butitsuremakesthingseasiertoread.”
Code is not write only.
Code is communication!
Code formatting and style
We have all written code which is hard to read!
We all improve over time.
The only way to write good code is to write tons of shitty code first. Feeling shame about bad code stops you from getting to good code
— Hadley Wickham (@hadleywickham) April 17, 2015
Code formatting and style
Some keys points:
- be consistent, emulate experienced coders
- use snake_case for variable names (not CamelCase, dot.case)
- use
<-(not=) for assignment
- use spacing around most operators and after commas
- use indentation
- avoid long lines, break up code blocks with new lines
- use
"for quoting text (not') unless the text contains double quotes
- space after
#for comments
😩 Ugly code 😩
data<-read_csv('../data-raw/Y101_Y102_Y201_Y202_Y101-5.csv',skip=2)
library(janitor);sol<-clean_names(data)
data=data|>filter(str_detect(description,"OS=Homo sapiens"))|>filter(x1pep=='x')
data=data|>
mutate(g=str_extract(description,
"GN=[^\\s]+")|>str_replace("GN=",''))
data<-data|>mutate(id=str_extract(accession,"1::[^;]+")|>str_replace("1::",""))😩 Ugly code 😩
- no spacing or indentation
- inconsistent splitting of code blocks over lines
- inconsistent use of quote characters
- no comments
- variable names convey no meaning
- use of
=for assignment and inconsistently - multiple commands on a line
- library statement in the middle of the analysis
😎 Cool code 😎
# Packages ----------------------------------------------------------------
library(tidyverse)
library(janitor)
# Import ------------------------------------------------------------------
# define file name
file <- "../data-raw/Y101_Y102_Y201_Y202_Y101-5.csv"
# import: column headers and data are from row 3
solu_protein <- read_csv(file, skip = 2) |>
clean_names()
# Tidy data ----------------------------------------------------------------
# filter out the bovine proteins and those proteins
# identified from fewer than 2 peptides
solu_protein <- solu_protein |>
filter(str_detect(description, "OS=Homo sapiens")) |>
filter(x1pep == "x")
# Extract the genename from description column to a column
# of its own
solu_protein <- solu_protein |>
mutate(genename = str_extract(description,"GN=[^\\s]+") |>
str_replace("GN=", ""))
# Extract the top protein identifier from accession column (first
# Uniprot ID after "1::") to a column of its own
solu_protein <- solu_protein |>
mutate(protid = str_extract(accession, "1::[^;]+") |>
str_replace("1::", ""))😎 Cool code 😎
library()calls collectedUses code sections to make it easier to navigate
Uses white space and proper indentation
Commented
Uses more informative name for the dataframe
Code ‘algorithmically’
Code ‘algorithmically’
Write code which expresses the structure of the problem/solution.
Avoid hard coding numbers if at all possible - declare variables instead
Declare frequently used values as variables at the start e.g., colour schemes, figure saving settings
😩 Hard coding numbers.
Suppose we want to calculate the sums of squares, \(SS(x)\), for the number of eggs in five nests.
The formula is given by: \(\sum (x_i- \bar{x})^2\)
We could calculate the mean and copy it, and the individual numbers into the formula
😩 Hard coding numbers.
# mean number of eggs per nest
sum(3, 5, 6, 7, 8) / 5[1] 5.8# ss(x) of number of eggs
(3 - 5.8)^2 + (5 - 5.8)^2 + (6 - 5.8)^2 + (7 - 5.8)^2 + (8 - 5.8)^2[1] 14.8I am coding the calculation of the mean rather using the mean() function only to explain what ‘coding algorithmically’ means using a simple example.
😩 Hard coding numbers
if any of the sample numbers must be altered, all the code needs changing
it is hard to tell that the output of the first line is a mean
its hard to recognise that the numbers in the mean calculation correspond to those in the next calculation
it is hard to tell that 5 is just the number of nests
no way of know if numbers are the same by coincidence or they refer to the same thing
😎 Better
😎 Better
the commenting is similar but it is easier to follow
if any of the sample numbers must be altered, only that number needs changing
assigning a value you will later use to a variable with a meaningful name allows us to understand the first and second calculations
makes use of R’s elementwise calculation which resembles the formula (i.e., is expressed as the general rule)
Naming things

Guiding principle - Have a convention! Good file names are:
machine readable
human readable
play nicely with sorting
Naming suggestions
no spaces in names
use snake_case or kebab-case rather than CamelCase or dot.case
use all lower case except very occasionally where convention is otherwise, e.g., README, LICENSE
ordering: use left-padded numbers e.g., 01, 02….99 or 001, 002….999
dates ISO 8601 format: 2020-10-16
write down your conventions
Workflow tips
multiple cursors
open a file/function or find a variable CONTROL+.
the command palette CONTROL+SHIFT+P
segmenting code CONTROL+SHIFT+R
to correct indentation CONTROL+i
to reformat code CONTROL+SHIFT+A Not perfect but corrects spacing, indentation, multiple commands on lines and assignment with
=to comment and uncomment lines CONTROL+SHIFT+C
Tools | Global options | Code | Display | Show margin
Tools | Global options | Code | Diagnostic | Provide R style diagnostics
It’s all gone wrong, Restart R CONTROL+SHIFT+F10
Summary
Use an RStudio project for any R work (you can also incorporate other languages)
Write Cool code not Ugly code: space, consistency, indentation, comments, meaningful variable names
Write code which expresses the structure of the problem/solution.
Avoid hard coding numbers if at all possible - declare variables instead
Reading
Completely optional suggestions for further reading
- Project-oriented workflow | What They Forgot to Teach You About R (Bryan et al., n.d.). Recommended if you still need convincing to use RStudio Projects
- Ten simple rules for reproducible computational research (Sandve et al. 2013)
- Good enough practices in scientific computing (Wilson et al. 2017)
- Excuse Me, Do You Have a Moment to Talk About Version Control? (Bryan 2018)
Pages made with R (R Core Team 2024), Quarto (Allaire et al. 2024), knitr (Xie 2024, 2015, 2014), kableExtra (Zhu 2021)