Independent Study to prepare for workshop
Transcriptomics 1: 👋 Hello data!
3 December, 2025
Overview
Concise summary of the experimental design and aims
What the raw data consist of
What has been done to the data so far
What steps we will take in the workshop
The Data
There are 3 transcriptomic datasets
🎄 bulk RNA-seq from Arabidopsis thaliana
💉 bulk RNA-seq from Leishmania mexicana
🐭 single cell RNA-seq from mouse stemcells
Experimental design
🎄 Experimental design

Schematic of arabidopsis experiment
🎄 Experimental design

Schematic of arabidopsis experiment
2 plant tissues
2 nickel conditions
6 replicates
2 x 2 x 6 = 24 samples
🎄 Experimental design

Schematic of arabidopsis experiment
2 plant tissues: root and aerial. This is the tissue treatment
2 nickel conditions: control and low Ni. This is the Ni treatment
6 replicates. These are the replicates
2 x 2 x 6 = 24 samples
🎄 Aim
Find genes that are “differentially expressed” between tissue types and nickel conditions e.g. root tissue grown under control and low Ni conditions
Differentially expressed means the expression in one group is significantly higher than in the other
🎄 Guided analysis
The workshops will take you through comparing the root tissue grown under control and low Ni conditions
You will make other comparisons independently
You will be guided to carefully document your work so you can apply the same methods to other comparisons
Do the independent study before and after the workshop!
💉 Experimental design
Schematic of leishmania experiment
💉 Experimental design

Schematic of leishmania experiment
3 stages
3 samples
3 x 3 = 9 samples
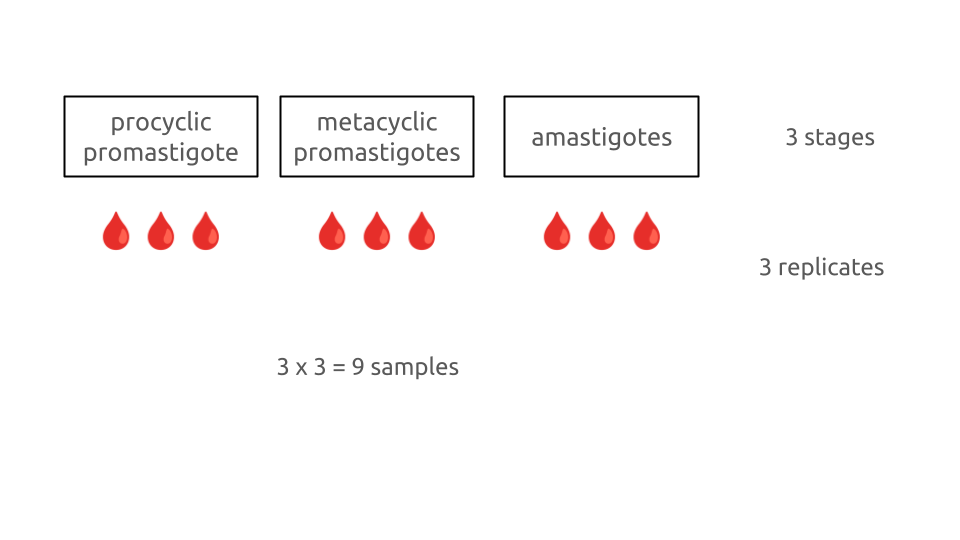
💉 Experimental design

Schematic of leishmania experiment
three stages: procyclic promastigotes, metacyclic promastigotes and amastigotes. This is the stage treatment
three samples. These are the replicates
3 x 3 = 9 samples
💉 Aim
Find genes that are “differentially expressed” between stages e.g., procyclic promastigotes and the metacyclic promastigotes
Differentially expressed means the expression in one group is significantly higher than in the other
💉 Guided analysis
The workshops will take you through comparing the procyclic promastigotes and the metacyclic promastigotes
You will make other comparisons independently
You will be guided to carefully document your work so you can apply the same methods to other comparisons
Do the independent study before and after the workshop!
🐭 Experimental design
Schematic of stem cell experiment
🐭 Experimental design

Schematic of stem cell experiment
Cells were sorted using flow cytometry on the basis of cell surface markers
There are 3 cell types
Many cells of each cell type were sequenced
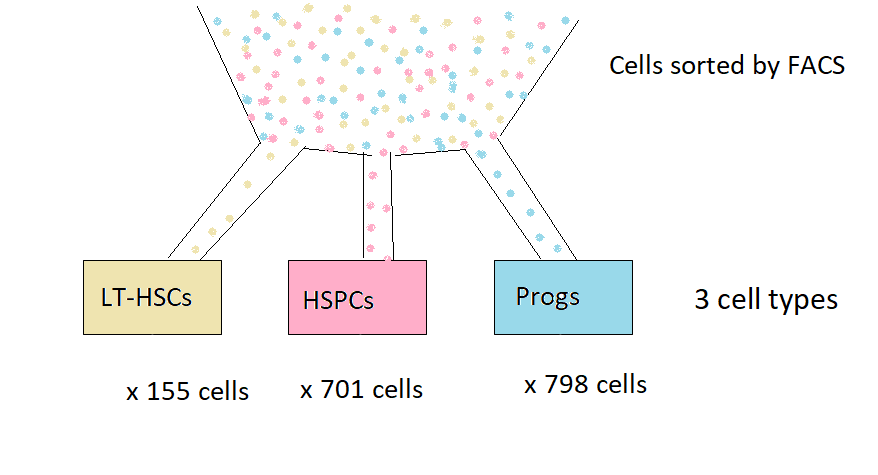
🐭 Experimental design

Schematic of stem cell experiment
There are three cell types: LT-HSCs, HSPCs, Progs This is the cell “treatment”
Many cells of each type were sequenced: These are the replicates
155 LT-HSCs, 701 HSPCs, 798 Progs
🐭 Aim
find genes that are “differentially expressed” between at least two cell types
Differentially expressed means the expression in one group is significantly higher than in the other
🐭 Guided analysis
The workshops will take you through comparing the HSPC and Prog cells
You will make other comparisons independently
You will be guided to carefully document your work so you can apply the same methods to other comparisons
Do the independent study before and after the workshop!
Where do the data come from?
Raw Sequence data
The raw data are “reads” from a sequencing machine in FASTQ files
A read is sequence of RNA which is shorter than the whole transcriptome
The length of the reads depends on the type of sequencing machine
- Short-read technologies (e.g. Illumina) have higher base accuracy but are harder to align
- Long-read technologies (e.g. Nanopore) have lower base accuracy but are easier to align
Raw Sequence data
Optional
You can read more about Sequencing technologies in Statistically useful experimental design(Rand and Forrester 2022)
What has been done to the data so far
General steps
Reads are filtered and trimmed on the basis of a quality score
They are then aligned/pseudo-aligned to a reference genome/transcriptome (or assembled de novo)
And then counted to quantify the expression
Counts need to be normalised to account for differences in sequencing depth and transcript length before, or as part of, statistical analysis.
🎄 Data
Brand spanking new! Provided by Alex Marks (Marks and Rylott, n.d.)
Expression for the whole transcriptome ENSEMBL Arabidopsis TAIR10(Yates et al. 2022)
Values are raw counts
The statistical analysis method we will use is
DESeq2(Love, Huber, and Anders 2014). It requires raw counts and performs the normalisation itself.
💉 Data
Brand spanking new!
Expression for the whole transcriptome L. mexicana MHOM/GT/2001/U1103(Rogers et al. 2011)
Values are raw counts
The statistical analysis method we will use is
DESeq2(Love, Huber, and Anders 2014). It requires raw counts and performs the normalisation itself.
🐭 Data
- Published in Nestorowa et al. (2016)
- Expression for a subset of genes, the secretome
Values are log2 normalised values
The statistical analysis method we will use is
scran(Lun, McCarthy, and Marioni 2016) and it requires normalised values
Workshops
Workshops
Transcriptomics 1: Hello data Getting to know the data. Checking the distributions of values overall, across rows and columns to check things are as we expect and detect rows/columns that need to be removed
Transcriptomics 2: Statistical Analysis. Identifying which genes are differentially expressed between treatments. This is the main analysis step. We will use different methods for bulk and single cell data.
Transcriptomics 3: Visualising. Principal Component Analysis (PCA) and volcano plots to visualise the results of the analysis.
References
Pages made with R (R Core Team 2024), Quarto (Allaire et al. 2024), knitr (Xie 2024, 2015, 2014), kableExtra (Zhu 2021)