install.packages("ggrepel")Independent Study to prepare for workshop
Transcriptomics 3: Visualising
3 December, 2025
Overview
In these slides we will:
Check where you are
-
learn some concepts used omics visualisation
- Principle Component Analysis (PCA)
- Volcano plots
Find out what packages to install before the workshop
Where should you be?
What we did in Transcriptomics 2: Statistical Analysis
carried out differential expression analysis
found genes not expressed at all, or expressed in one group only
Saved results files
Where should you be?
After the Transcriptomics 2: 👋 Statistical Analysis Workshop including:
the Independent Study to consolidate, you should have:
🎄 Arabidopisis
An RStudio Project called arab-88H which contains:
- data-raw:
arabidopsis-root.csv,arabidopsis-aerial.csv - data-processed:
root_filtered.csv,aerial_filtered.csv - results:
root_cont_only.csv,root_lowni_only.csv,root_results.csv, and equivalents for aerial - Two scripts:
cont-low-root.R,cont-low-aerial.R
💉 Leishmania
An RStudio Project called leish-88H which contains:
- data-raw:
leishmania-mex-ama.csv,leishmania-mex-pro.csv,leishmania-mex-meta.csv - data-processed:
pro_meta_filtered.csv,pro_ama_filtered.csv - results:
pro_meta_results.csv,pro_ama_results.csv - Two scripts:
pro_meta.R,pro_ama.R
🐭 Stem cells
An RStudio Project called mice-88H which contains:
- data-raw:
secretome_hspc.csv,secretome_prog.csv,secretome_lthsc.csv
- data-processed:
hspc_prog.csv,hspc_lthsc.csv - results:
hspc_prog_results.csv,hspc-lthsc_results.csv, - Two scripts:
hspc-prog.R,hspc-lthsc.R
Additionally…
Files should be organised into folders. Code should well commented and easy to read. You should have curated your code to remove unnecessary commands that were useful to troubleshoot or understand objects in your environment but which are not needed for the final analysis.
If you are missing files, go through:
Go through:
Examine the results files
All results files
Remind yourself of the key columns in any of the results files:
- normalised counts for each sample/cell
- a log2 fold change
- an unadjusted p-value
- a p value adjusted for multiple testing (called
FDRorpadj) - a gene id
- other information about each gene
🎄 , 💉 results files
-
baseMeanis the mean of the normalised counts for the gene across all samples -
lfcSEstandard error of the fold change -
statis the test statistic (the Wald statistic)
🐭 Stem cells
- Top is the rank of the gene ordered by the p-value (smallest first)
-
summary.logFCandlogFC.hspcgive the same value (in this case since comparing two cell types)
Plots
What is the purpose of a Transcriptomics plot?
In general, we plot data to help us summarise and understand it
This is especially import for transcriptomics data where we have a very large number of variables and often a large number of observations
We will look at two plots very commonly used in transcriptomics analysis: Principal Component Analysis (PCA) plot and Volcano Plots
Principal Component Analysis (PCA)
PCA
Principal Component Analysis is an unsupervised machine learning technique
Unsupervised methods1 are unsupervised in that they do not use/optimise to a particular output. The goal is to uncover structure. They do not test hypotheses
It is often used to visualise high dimensional data because it is a dimension reduction technique
PCA
Takes a large number of continuous variables (like gene expression) and reduces them to a smaller number of variables (called principal components) that explain most of the variation in the data
The principal components can be plotted to see how samples cluster together
PCA
- To understand the logic of PCA, imagine we might plot the expression of one gene against that of another
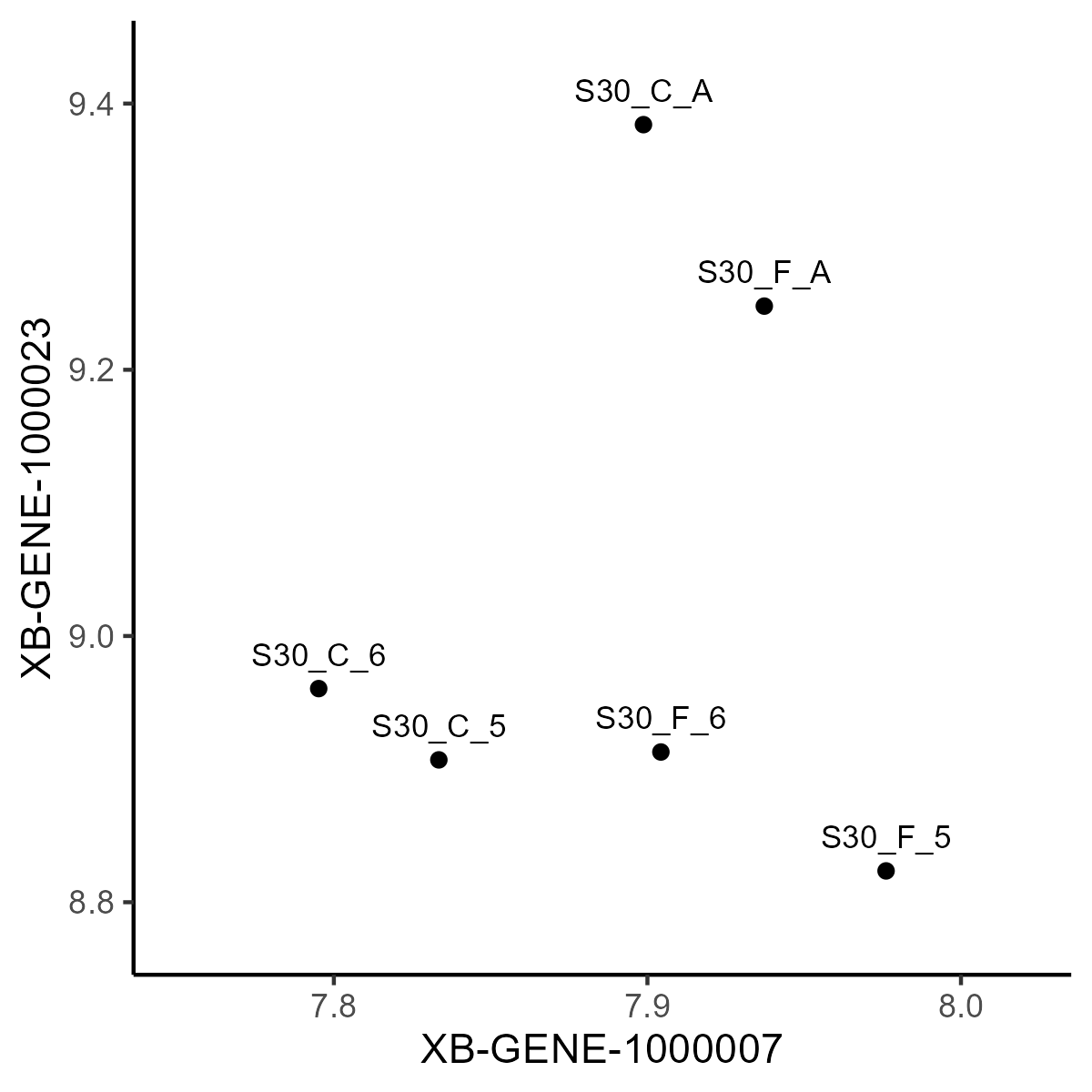

This gives us some in insight in how the sample/cells cluster. But we have a lot of genes (even for the stem cells) to consider. How do we know if the pair we use is typical? How can we consider all the genes at once?
PCA
- PCA is a solution for this - It takes a large number of continuous variables (like gene expression) and reduces them to a smaller number of “principal components” that explain most of the variation in the data.
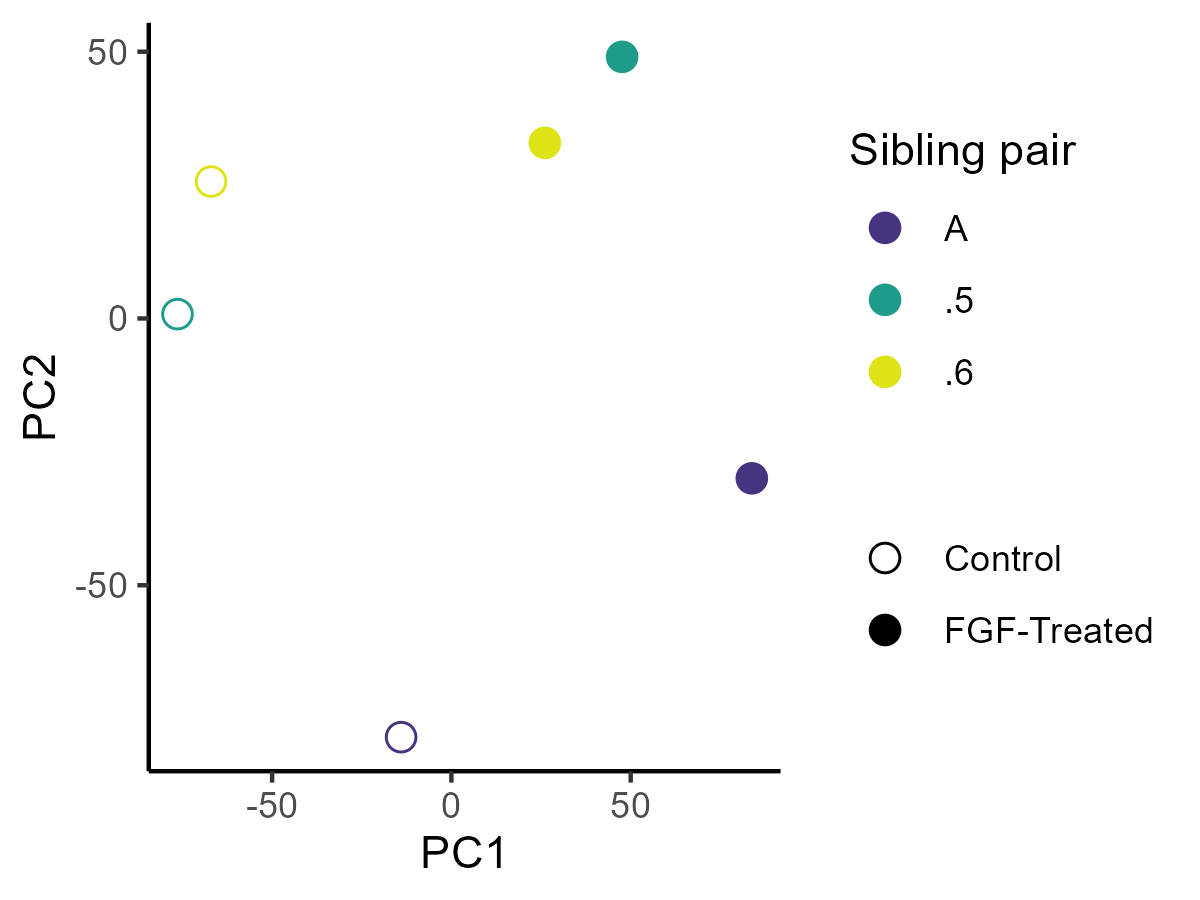
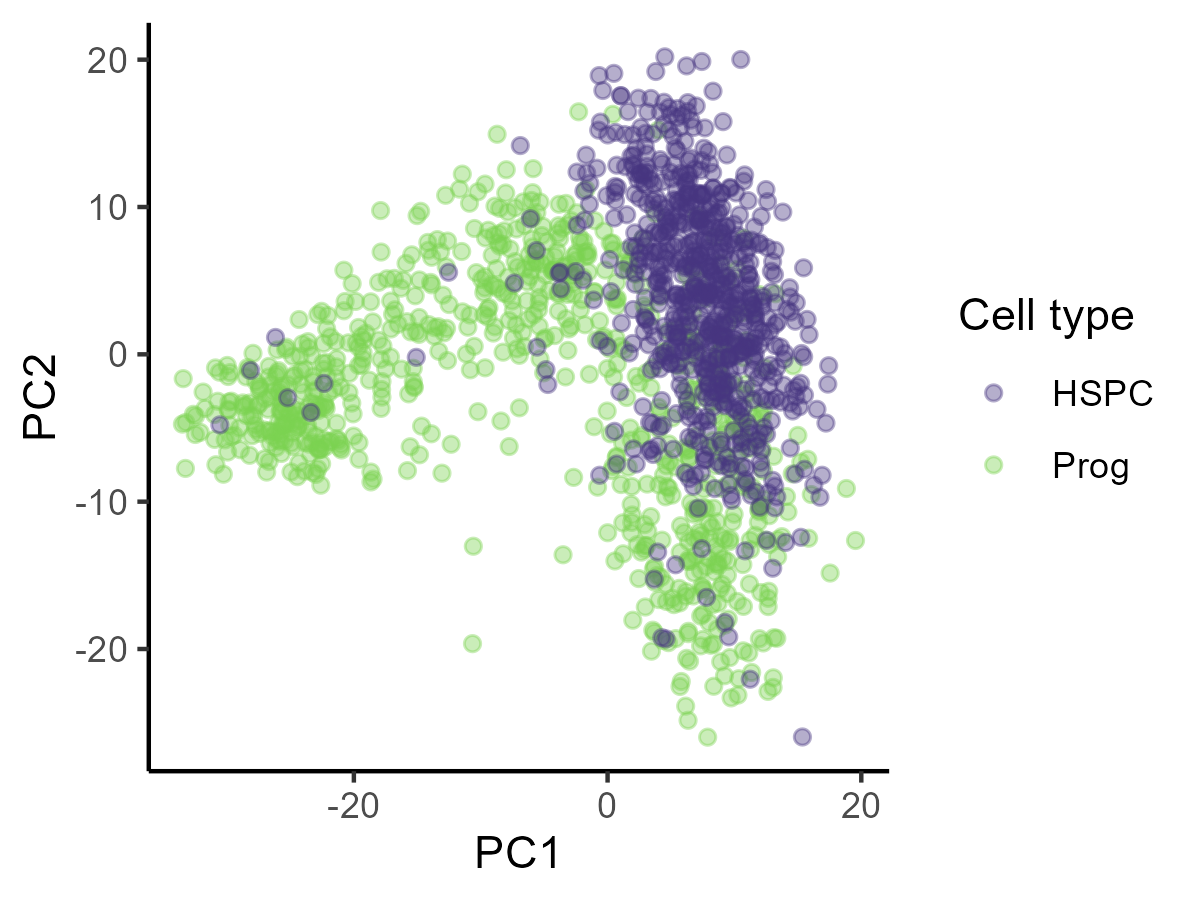
PCA
We have done PCA after differential expression, but often PCA might is one of the first exploratory steps because it gives you an idea whether you expect general patterns in gene expression that distinguish groups.
Volcano plots
Volcano plots
Volcano plots often used to visualise the results of differential expression analysis
They are just a scatter of the adjusted p value against the fold change….
almost - in fact, we plot the negative log of the adjusted p-value against the log fold change
Why?
Volcano plots
It is because small probabilities are important, large ones are not which means the axis is counter intuitive because small p-values (i.e., significant values) are at the bottom of the axis)
And since p-values range from 1 to very tiny the important points are all squashed at the bottom of the axis
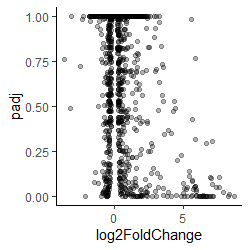
Volcano plot padj against fold change
Volcano plots
- By plotting the negative log of the adjusted p-value the values are spread out, and the most significant are at the top of the axis
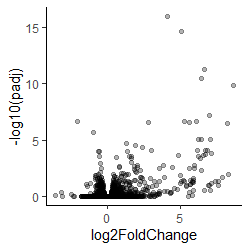
Volcano plot -log(adjusted p) against fold change
Visualisations
Should be done on normalised data so meaningful comparisons can be made
The 🐭 stem cell data were already log2normalised
The other datasets were normalised by the DE method and we saved the values to the results files. We will log transform them in the workshop
Packages
This package is on the University computers which you can access on campus or remotely using the VDS
If you want to use your own machine you will need to install the package.
Install ggrepel from CRAN in the the normal way:
This package allows you to label points on a plot without them overlapping.
Workshops
Workshops
Transcriptomics 1: Hello data Getting to know the data. Checking the distributions of values overall, across rows and columns to check things are as we expect and detect rows/columns that need to be removed
Transcriptomics 2: Statistical Analysis. Identifying which genes are differentially expressed between treatments. This is the main analysis step. We will use different methods for bulk and single cell data.
Transcriptomics 3: Visualising. Principal Component Analysis (PCA) volcano plots to visualise the results of the