📦
You can make an R package too!
UoY, Bioinformatics group
Emma Rand
Summary
- Why make a package?
- Where packages come from and where do they live?
- Package States
- How to make a minimal documented package and check it using the
devtools(Wickham et al. 2021) approach - Components of a minimal package
Prerequisites
You should have
- R and RStudio
- R build toolchain: Rtools(windows) or XCode (mac) or r-base-dev
-
devtoolsandassertthat - optionally git, a GitHub account and verified it can talk to RStudio
Learning Objectives
by the end of this session you will able to:
- explain the rationale for writing packages
- describe the different states a package can be in
- create a minimal package and explain its key components
- add functions with
usethis:use_r() - use the package interactively with
devtools::load_all() - use
devtools::check()to executeR CMDcheck - add a license with
usethis::use_mit_license() - add documentation using
roxygen2anddevtools::document() - add package dependencies with
usethis::use_package()
Why make a package?
Why make a package?
Conventionally:
- Package developers
- Generalisable analytical methods
- For use on other data
- Public release
Why make a package?
Package up your data analysis project!
- You don’t need to have a collection of highly generalisation functions
- You don’t need to share it with anyone else
- If you are already trying to work reproducibly you are almost doing it anyway!
- But it will help you do it better
Why make a package?
If you are already trying to work reproducibly you are almost doing it anyway!
/stem-cell-proteomic
/data-raw
/data-processed
/figures
/R
README.md
report.Rmd
stem-cell-proteomic.RProj… making a package is just a short step beyond that
Script vs package
Script
Package
- defines reusable components
- defined by presence of
DESCRIPTIONfile - Required packages specified in
DESCRIPTION, made available inNAMESPACEfile - documentation in files and
Roxygencomments - Install and restart
Be nice to future you
Future self: CC-BY-NC, by Julen Colomb, derived from Randall Munroe cartoon
To avoid
Where packages come from and live?
Where do R packages come from?
CRAN:
GitHub:
Bioconductor
Where do packages live?
In a library! In
The R home directory is the top-level directory of your R installation.
Note: this is not the same as your working directory or your home directory.
Your R installation
Your library
Your library
[1] "askpass" "assertthat" "backports" "base"
[5] "base64enc" "bit" "bit64" "blob"
[9] "boot" "brew" "brio" "broom"
[13] "bslib" "cachem" "callr" "cellranger"
[17] "class" "cli" "clipr" "cluster"
[21] "codetools" "colorspace" "commonmark" "compiler"
[25] "cpp11" "crayon" "credentials" "curl"
[29] "data.table" "datasets" "DBI" "dbplyr"
[33] "desc" "devtools" "diffobj" "digest"
[37] "downlit" "dplyr" "dtplyr" "ellipsis"
[41] "evaluate" "fansi" "farver" "fastmap"
[45] "forcats" "foreign" "fs" "gargle"
[49] "generics" "gert" "ggplot2" "gh"
[53] "gitcreds" "glue" "googledrive" "googlesheets4"
[57] "graphics" "grDevices" "grid" "gtable"
[61] "haven" "highr" "hms" "htmltools"
[65] "httr" "ids" "ini" "isoband"
[69] "jquerylib" "jsonlite" "KernSmooth" "knitr"
[73] "labeling" "lattice" "lifecycle" "lubridate"
[77] "magrittr" "MASS" "Matrix" "memoise"
[81] "methods" "mgcv" "mime" "modelr"
[85] "munsell" "nlme" "nnet" "openssl"
[89] "parallel" "pillar" "pkgbuild" "pkgconfig"
[93] "pkgload" "praise" "prettyunits" "processx"
[97] "progress" "ps" "purrr" "R6"
[101] "rappdirs" "rcmdcheck" "RColorBrewer" "readr"
[105] "readxl" "rematch" "rematch2" "remotes"
[109] "reprex" "rlang" "rmarkdown" "roxygen2"
[113] "rpart" "rprojroot" "rstudioapi" "rversions"
[117] "rvest" "sass" "scales" "selectr"
[121] "sessioninfo" "spatial" "splines" "stats"
[125] "stats4" "stringi" "stringr" "survival"
[129] "sys" "tcltk" "testthat" "tibble"
[133] "tidyr" "tidyselect" "tidyverse" "tinytex"
[137] "tools" "translations" "tzdb" "usethis"
[141] "utf8" "utils" "uuid" "vctrs"
[145] "viridisLite" "vroom" "waldo" "whisker"
[149] "withr" "xfun" "xml2" "xopen"
[153] "yaml" "zip" Package states
Package states
There are five states a package can be in:
source
bundled
binary
installed
in-memory
Package states
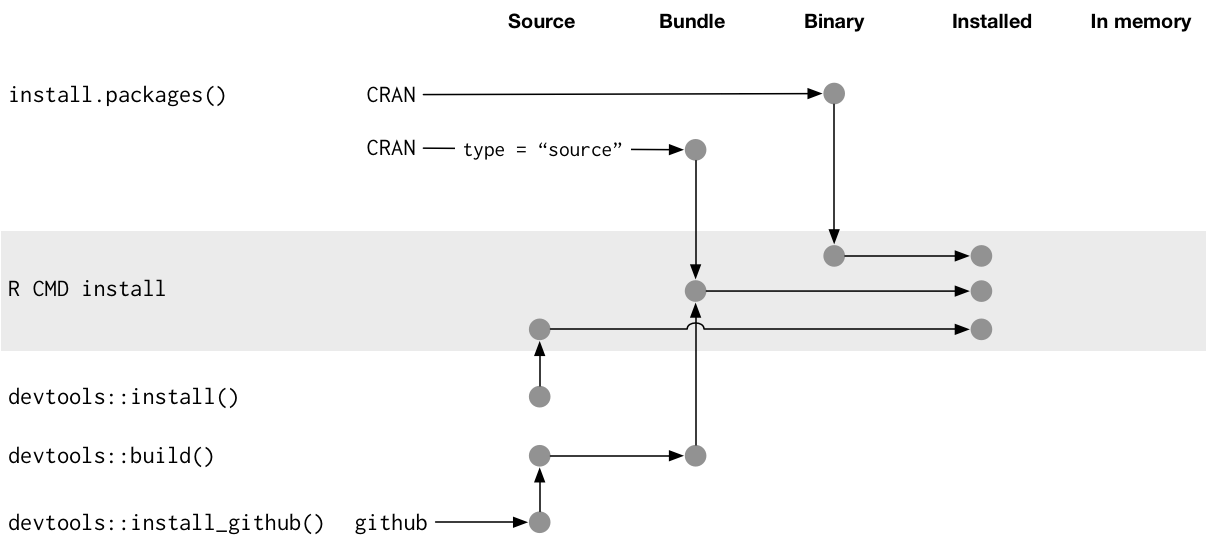
Package states
source
bundled
binary
installed
in-memory
What you create and work on.
Specific directory structure with some particular components e.g., DESCRIPTION, an R/ directory.
Package states
source
bundled
binary
installed
in-memory
Also known as “source tarballs”.
Package files compressed to single file.
Conventionally .tar.gz
You don’t normally need to make one.
Unpacked it looks very like the source package
Package states
source
bundled
binary
installed
in-memory
Package distribution for users w/o dev tools
Also a single file
Platform specific: .tgz (Mac) .zip (Windows)
Package developers submit a bundle to CRAN; CRAN makes and distributes binaries
Package states
source
bundled
binary
installed
in-memory
A binary package that’s been decompressed into a package library
Command line tool R CMD INSTALL powers all package installation
Package states
source
bundled
binary
installed
in-memory
If a package is installed, library() makes its function available by loading the package into memory and attaching it to the search path.
We do not use library() for packages we are working on
devtools::load_all() loads a source package directly into memory.
Create a package!
Create a package
Be deliberate about where you create your package
Do not nest inside another RStudio project, R package or git repo.
🎬 Create a package:
√ Creating 'C:/Users/er13/Desktop/mypackage/'
√ Setting active project to 'C:/Users/er13/Desktop/mypackage'
√ Creating 'R/'
√ Writing 'DESCRIPTION'
Package: mypackage
Title: What the Package Does (One Line, Title Case)
Version: 0.0.0.9000
Authors\@R (parsed):
* First Last \<first.last\@example.com\> \[aut, cre\] (YOUR-ORCID-ID)
Description: What the package does (one paragraph).
License: \`use_mit_license()\`, \`use_gpl3_license()\` or friends to
pick a license
Encoding: UTF-8
LazyData: true
Roxygen: list(markdown = TRUE)
RoxygenNote: 7.1.1
√ Writing 'NAMESPACE'
√ Writing 'mypackage.Rproj'
√ Adding '.Rproj.user' to '.gitignore'
√ Adding '\^mypackage\\\\.Rproj\$', '\^\\\\.Rproj\\\\.user\$' to '.Rbuildignore'
√ Opening 'C:/Users/er13/Desktop/mypackage/' in new RStudio session
√ Setting active project to '\<no active project\>'create_package()
What happens when we run create_package()?
R will create a folder called
mypackagewhich is a package and an RStudio projectrestart R in the new project
create some infrastructure for your package
start the RStudio Build pane
create_package()
What happens when we run create_package()?
mypackage.Rprojis the file that makes this directory an RStudio Project.DESCRIPTIONprovides metadata about your package.The
R/directory is where we will put.Rfiles with function definitions.NAMESPACEdeclares the functions your package exports for external use and the external functions your package imports from other packages.
create_package()
What happens when we run create_package()?
.Rbuildignorelists files that we need but that should not be included when building the R package from source..gitignoreanticipates Git usage and ignores some standard, behind-the-scenes files created by R and RStudio.
Add a function
Functions will go in an .R file.
There’s a usethis helper for adding .R files!
usethis::use_r() adds the file extension and saves in R/ folder
usethis::use_r()
🎬 Create a new R file in your package called animal_sounds.R
Add the function
🎬 Put the following code into your script:
Test your function
Development workflow
In a normal script you might use:
but when building packages we use a devtools approach
Development workflow
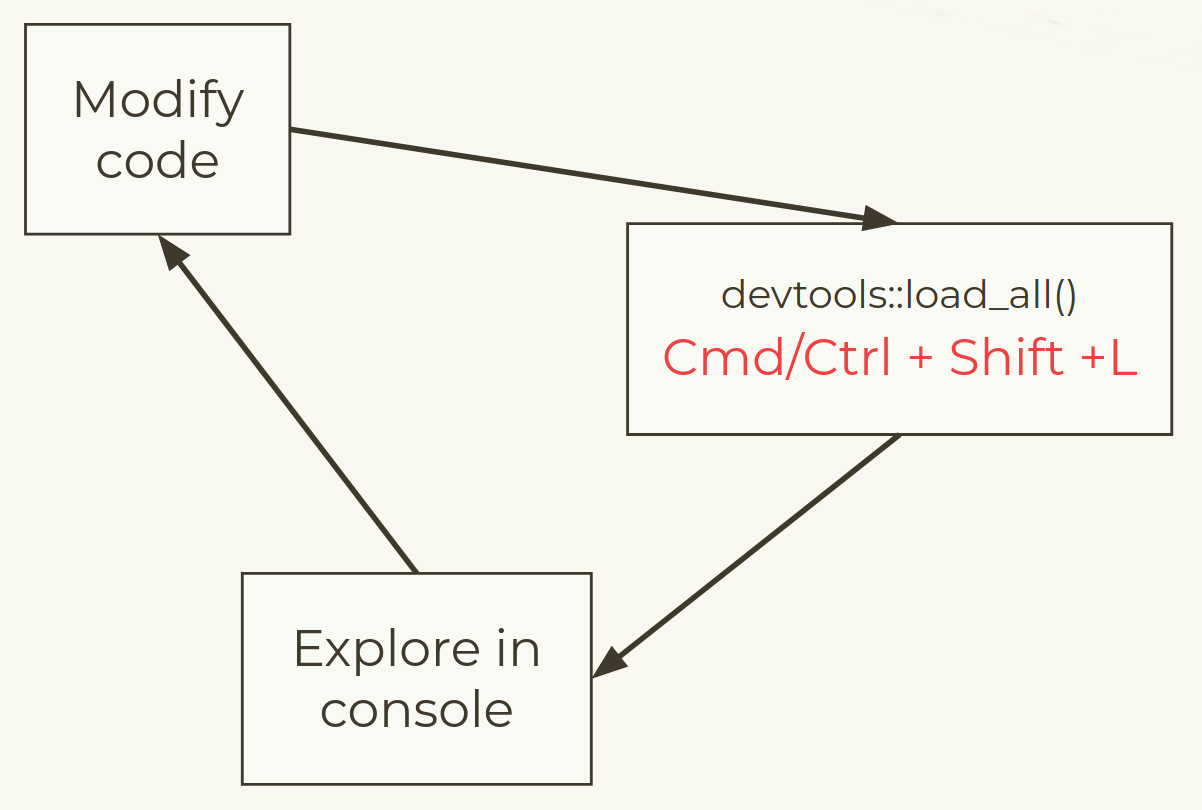
Development workflow
Load your package
devtools::load_all()
🎬 Load package with devtools::load_all().
Loading mypackageTest
Test the animal_sounds() function in the console.
[1] "The dog goes woof!"devtools::load_all()
🎬 Change some tiny thing about your function - maybe the animal “says” instead of “goes”?
🎬 Load with devtools::load_all() and test the updated function.
Check your package
Check your package
R CMD check is the gold standard for checking that an R package is in full working order.
It is a programme that is executed in the shell.
However, devtools has the check() function to allow you to run this without leaving your R session.
devtools::check()
🎬 Check your package
devtools::check()
You will get lots of output ending with:
-- R CMD check results -------------------- mypackage 0.0.0.9000 ----
Duration: 12.5s
> checking DESCRIPTION meta-information ... WARNING
Non-standard license specification:
`use_mit_license()`, `use_gpl3_license()` or friends to pick a license
Standardizable: FALSE
> checking dependencies in R code ... WARNING
'::' or ':::' import not declared from: 'assertthat'
0 errors √ | 2 warnings x | 0 notes √Aside: in case of error
On running devtools::check() you may get an error if you are using a networked drive.
Updating mypackage documentation
Error: The specified file is not readable: path-to\mypackage\NAMESPACEThis is covered here and can be fixed.
Aside: in case of error
Save a copy of this file:
Save it somewhere other than the mypackage directory
Open the file from the mypackage project session
Run the whole file
You should now find that devtools::check() proceeds normally
License
Add a license
usethis helps out again! use_mit_license(), use_agpl_license(), use_ccby_license() etc
🎬 Add a MIT license1 - use your own name!
🎬 What files have appeared?
🎬 How has the DESCRIPTION file changed?
🎬 Run devtools::check() again. Has one of the warnings disappeared?
Document your package
Levels of package documentation
Metadata: The
DESCRIPTIONfile – an overview of “what’s in this package?”Object documentation: Documentation for each of the exported functions and datasets in the package, along with examples of usage
Vignettes: Long form documentation, generally discussing how to use a number of functions from the - package together and/or how a package fits into a larger ecosystem of packages
pkgdown sites: Websites for your package!
Metadata in DESCRIPTION
Title: One line, title case, with no period. Fewer than 65 characters.
-
Version
- for release: MAJOR.MINOR.PATCH version.
- for development version building on version MAJOR.MINOR.PATCH, use: MAJOR.MINOR.PATCH.9000
Authors@R: “aut” means author, “cre” means creator, “ctb” means contributor.
Description: One paragraph describing what the package does. Keep the width of the paragraph to 80 characters; indent subsequent lines with 4 spaces.
License
Encoding: How to encode text, use UTF-8 encoding.
LazyData: Use true to lazy-load data sets in the package.
Update DESCRIPTION
🎬 Add a title and description.
🎬 Add yourself as an author and creator.
Object documentation
Object documentation is what you see when you use ? or help() to find out more about a function or a dataset in a package.
We will create object documentation using Roxygen comments, which start with #’
Much of the work is done by the roxygen2 package, but we won’t directly run roxygen2 functions, instead run functions from devtools that call them
Object documentation workflow
Add roxygen comments to your .R files.
Run
devtools::document()to convert roxygen comments to .Rd files.Load the current version of the package with
devtools::load_all()Preview documentation with
?Rinse and repeat until the documentation looks the way you want.
Document your function
🎬 Open animal_sounds.R
🎬 Go to Code > Insert Roxygen Documentation
🎬 Fill in the documentation: Give your function a title, then, in a new paragraph, a brief description, define the two parameters, and finally, describe what the function returns
🎬 Save animal_sounds.R, run devtools::document() followed by devtools::load_all()
🎬 Preview the documentation with ?animal_sounds and edit your documentation if anything needs to be changed
Add examples
🎬 Under @examples, add one example for using your function
🎬 Save animal_sounds.R, run devtools::document() followed by devtools::load_all()
🎬 Preview the documentation with ?animal_sounds and edit your documentation if anything needs to be changed
Package dependencies
Remember this?
We have used a function from the
assertthatpackage in a function in our packageBut we haven’t declared that officially, we need to do that
Package dependencies
- Imports: Packages listed here must be installed for your package to work. If they’re not, they will get installed along with the package you’re installing.
- Suggests: Packages listed here are used by your package, but not required for your package. You might use suggested packages for example datasets, to run tests, build vignettes, or maybe there’s only one function that needs the package.
- Depends: Avoid where possible, but you might use it to require a specific version of R, e.g. Depends: R (>= 3.4.0). Think critically before doing this as it will have downstream effects on other packages that depend on your package.
Package Imports
usethis::use_package() is there for us again! It defaults to imports1
🎬 Use usethis::use_package() to add the assertthat package to Imports
Package Imports
🎬 How your DESCRIPTION file changed?
🎬 Run devtools::check() again. Has the warning disappeared?
📦 Woo hoo📦
You made a package!
Licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. 
Summary
- It is useful to make a package
- it is fairly easy with
devtools - it will help you work more reproducibly
- you don’t have to share
- it is fairly easy with
- Packages can be in one of 5 states:
- source - what you write
- bundled - source compressed to single file, submitted to CRAN
- binary - distribution for users w/o
devtools - installed - a binary that’s been decompressed
- in-memory - installed package that has been loaded
Summary continued
- A minimal package comprises
- a folder which is a package and a RProj
-
DESCRIPTION,NAMESPACE,.Rbuildignore.gitignore -
R/directory for functions
- We add functions with
usethis:use_r() - We use the package interactively with
devtools::load_all() - We use
devtools::check()to executeR CMDcheck - We add a license with
usethis::use_mit_license() - We add documentation using
roxygen2anddevtools::document() - We add package dependencies with
usethis::use_package()