📦
You can make an R package too!
Summary
- Why make a package?
- Where packages come from and where do they live?
- Package States
- How to make a minimal documented package and check it using the
devtools(Wickham et al. 2022) approach - Components of a minimal package
Prerequisites
You should have
- R and RStudio
- R build toolchain: Rtools(windows) or XCode (mac) or r-base-dev
-
devtoolsandassertthat
Learning Objectives
By the end of this session you will able to:
- explain the rationale for writing packages
- describe the different states a package can be in
- create a minimal package and explain its key components
- add functions with
usethis:use_r() - use the package interactively with
devtools::load_all() - use
devtools::check()to executeR CMD check - add a license with
usethis::use_mit_license() - add documentation using
roxygen2anddevtools::document() - add package dependencies with
usethis::use_package()
Why make a package?
Why make a package?
Conventionally:
- Package developers
- Generalisable analytical methods
- For use on other data
- Public release
Why make a package?
But you can also make a package just for your own use! Or just for a data analysis project!
- You don’t need to have a collection of highly generalisable functions
- You don’t need to share it with anyone else
- If you are already trying to work reproducibly you are almost doing it anyway!
- But it will help you do it better
Why make a package?
If you are already trying to work reproducibly you are almost doing it anyway!
/stem-cell-proteomic
/data-raw
/data-processed
/figures
/R
README.md
report.Rmd
stem-cell-proteomic.RProj… making a package is just a short step beyond that
Be nice to future you
Future self: CC-BY-NC, by Julen Colomb, derived from Randall Munroe cartoon
To avoid
Where packages come from and live?
Where do R packages come from?
CRAN:
GitHub:
Bioconductor
Where do packages live?
In a library! In
[1] "C:/Users/er13/AppData/Local/R/win-library/4.5"
[2] "C:/Program Files/R/R-4.5.2/library" A library is a folder
Base packages live in C:/Program Files/R/R-4.5.2/library
Package states
Package states
There are five states a package can be in:
source
bundled
binary
installed
in-memory
Package states
source
bundled
binary
installed
in-memory
What you write.
Specific directory structure and components e.g., DESCRIPTION, an R/ directory.
Package states
source
bundled
binary
installed
in-memory
Package files compressed to single file.
Conventionally .tar.gz
Package states
source
bundled
binary
installed
in-memory
Standard package distribution
Platform specific: .tgz (Mac) .zip (Windows)
Package developers submit a bundle to CRAN; CRAN makes and distributes binaries
Package states
source
bundled
binary
installed
in-memory
A binary package that’s been decompressed into a package library
Package states
source
bundled
binary
installed
in-memory
If a package is installed, library() makes its functions available by loading the package into memory and attaching it to the search path.
Not used when making a package; devtools::load_all() instead.
Create a package!
Create a package
Be deliberate about where you create your package
Do not nest inside another RStudio project, R package or git repo.
🎬 Create a package:
√ Creating 'C:/Users/er13/Desktop/mypackage/'
√ Setting active project to 'C:/Users/er13/Desktop/mypackage'
√ Creating 'R/'
√ Writing 'DESCRIPTION'
Package: mypackage
Title: What the Package Does (One Line, Title Case)
Version: 0.0.0.9000
Authors\@R (parsed):
* First Last \<first.last\@example.com\> \[aut, cre\] (YOUR-ORCID-ID)
Description: What the package does (one paragraph).
License: \`use_mit_license()\`, \`use_gpl3_license()\` or friends to
pick a license
Encoding: UTF-8
LazyData: true
Roxygen: list(markdown = TRUE)
RoxygenNote: 7.1.1
√ Writing 'NAMESPACE'
√ Writing 'mypackage.Rproj'
√ Adding '.Rproj.user' to '.gitignore'
√ Adding '\^mypackage\\\\.Rproj\$', '\^\\\\.Rproj\\\\.user\$' to '.Rbuildignore'
√ Opening 'C:/Users/er13/Desktop/mypackage/' in new RStudio session
√ Setting active project to '\<no active project\>'create_package()
What happens when we run create_package()?
R will create a folder called
mypackagewhich is a package and an RStudio projectrestart R in the new project
create some infrastructure for your package
start the RStudio Build pane
create_package()
What happens when we run create_package()?
mypackage.Rprojmakes this directory an RStudio Project.DESCRIPTIONprovides metadata about your package.The
R/directory for.Rfiles with function definitions.NAMESPACEdeclares the functions your package exports and the functions it imports from other packages.
create_package()
What happens when we run create_package()?
.Rbuildignorelists files that we need but that should not be included when building the R package from source..gitignoreanticipates Git usage and ignores some standard, behind-the-scenes files created by R and RStudio.
Add a function
Functions will go in an .R file.
There’s a usethis helper for adding .R files!
usethis::use_r() adds the file extension and saves in R/ folder
usethis::use_r()
🎬 Create a new R file in your package called animal_sounds.R
Add the function
🎬 Put the following code into your script:
Test your function
Development workflow
In a normal script you might use:
but when building packages we use a devtools approach
Development workflow
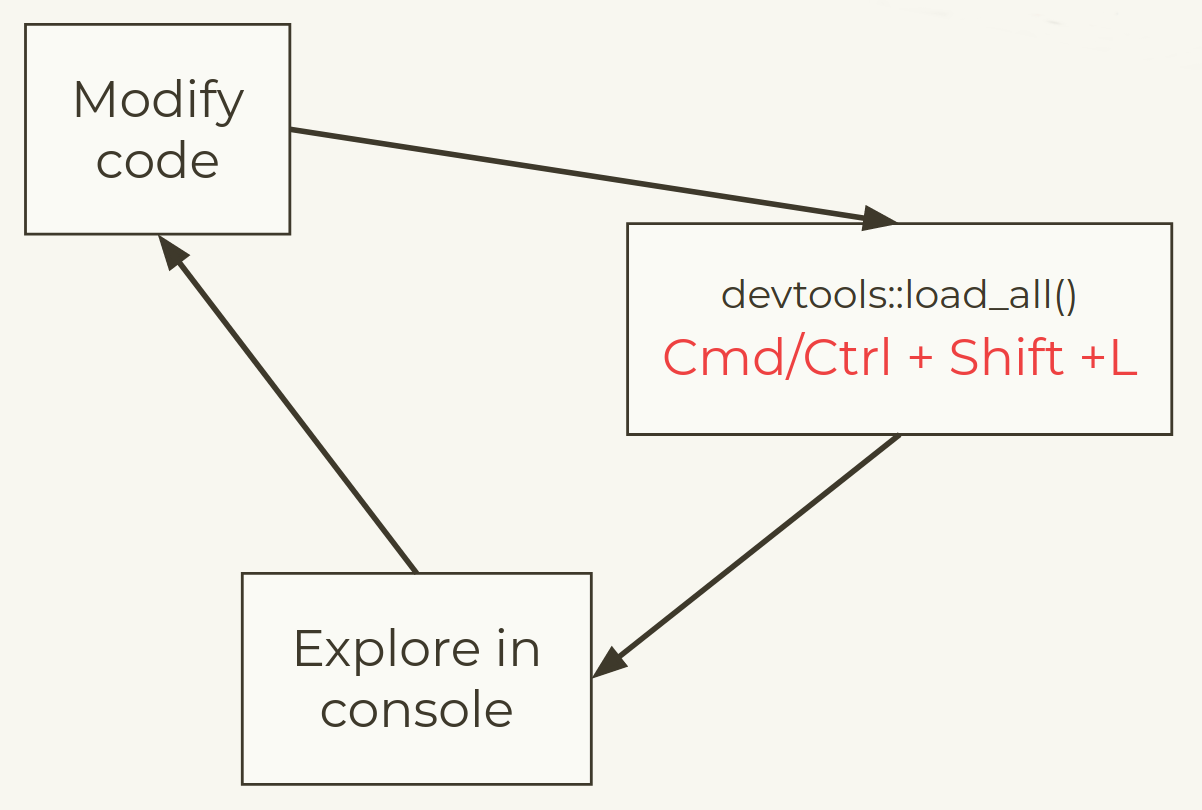
Development workflow
Load your package
devtools::load_all()
🎬 Load package with devtools::load_all().
Loading mypackageTest
Test the animal_sounds() function in the console.
[1] "The dog goes woof!"devtools::load_all()
🎬 Change some tiny thing about your function - maybe the animal “says” instead of “goes”?
🎬 Load with devtools::load_all() and test the updated function.
Check your package
Check your package
R CMD check is the gold standard for checking that an R package is in full working order.
It is a programme that is executed in the shell.
However, devtools has the check() function to allow you to run this without leaving your R session.
devtools::check()
🎬 Check your package
devtools::check()
You will get lots of output ending with:
── R CMD check results ───────────────────────────────────── mypackage 0.0.0.9000 ────
Duration: 19.5s
❯ checking dependencies in R code ... WARNING
'::' or ':::' import not declared from: 'assertthat'
❯ checking DESCRIPTION meta-information ... NOTE
Invalid licence file pointers: LICENSE
0 errors ✔ | 1 warning ✖ | 1 note ✖License
Add a license
usethis helps out again! use_mit_license(), use_agpl_license(), use_ccby_license() etc
🎬 Add a MIT license1 - use your own name!
🎬 What files have appeared?
🎬 How has the DESCRIPTION file changed?
🎬 Run devtools::check() again. Has one of the warnings disappeared?
Document your package
Levels of package documentation
Metadata: The
DESCRIPTIONfile – an overview of “what’s in this package?”Object documentation: Documentation for each of the exported functions and datasets in the package, along with examples of usage
Vignettes: Long form documentation, generally discussing how to use a number of functions from the - package together and/or how a package fits into a larger ecosystem of packages
pkgdown sites: Websites for your package!
Metadata in DESCRIPTION
Title: One line, title case, with no period. Fewer than 65 characters.
-
Version
- for release: MAJOR.MINOR.PATCH version.
- for development version building on version MAJOR.MINOR.PATCH, use: MAJOR.MINOR.PATCH.9000
Authors@R: “aut” means author, “cre” means creator, “ctb” means contributor.
Description: One paragraph describing what the package does. Keep the width of the paragraph to 80 characters; indent subsequent lines with 4 spaces.
License
Encoding: How to encode text, use UTF-8 encoding.
LazyData: Use true to lazy-load data sets in the package.
Update DESCRIPTION
🎬 Add a title and description.
🎬 Add yourself as an author and creator.
Object documentation
Object documentation is what you see when you use ? or help() to find out more about a function or a dataset in a package.
Created by adding “Roxygen comments” to the function (\#')
Much of the work is done by the roxygen2 package called by ‘devtools’
Object documentation workflow
Add roxygen comments to your .R files.
Run
devtools::document()to convert roxygen comments to .Rd files.Load the current version of the package with
devtools::load_all()Preview documentation with
?Rinse and repeat until the documentation looks the way you want.
Document your function
🎬 Open animal_sounds.R
🎬 Go to Code > Insert Roxygen Documentation
🎬 Fill in the documentation: Title, a brief description, define the two parameters, and finally, describe what the function returns
🎬 Save animal_sounds.R, run devtools::document() followed by devtools::load_all()
🎬 Preview the documentation with ?animal_sounds and edit your documentation if anything needs to be changed
Add examples
🎬 Under @examples, add one example for using your function
🎬 Save animal_sounds.R, run devtools::document() followed by devtools::load_all()
🎬 Preview the documentation with ?animal_sounds and edit your documentation if anything needs to be changed
Package dependencies
Remember this?
We have used a function from the
assertthatpackage in a function in our packageBut we haven’t declared that officially, we need to do that
Types of dependency
- Imports: must be installed for your package to work.
- Suggests: used but not required for your package. e.g., datasets, for tests or vignettes, or limited use.
- Depends: Avoid where possible, but you might use it to require a specific version of R, e.g. Depends: R (>= 3.4.0).
Package Imports
usethis::use_package() is there for us again! It defaults to imports1
🎬 Use usethis::use_package() to add the assertthat package to Imports
Package Imports
🎬 How your DESCRIPTION file changed?
🎬 Run devtools::check() again. Has the warning disappeared?
📦 Woo hoo📦
You made a package!
Licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. 
Summary
- It is useful to make a package
- it is fairly easy with
devtools - it will help you work more reproducibly
- you don’t have to share
- it is fairly easy with
- Packages can be in one of 5 states:
- source - what you write
- bundled - source compressed to single file, submitted to CRAN
- binary - distribution for users w/o
devtools - installed - a binary that’s been decompressed
- in-memory - installed package that has been loaded
Summary continued
- A minimal package comprises
- a folder which is a package and a RProj
-
DESCRIPTION,NAMESPACE,.Rbuildignore.gitignore -
R/directory for functions
- We add functions with
usethis:use_r() - We use the package interactively with
devtools::load_all() - We use
devtools::check()to executeR CMD check - We add a license with
usethis::use_mit_license() - We add documentation using
roxygen2anddevtools::document() - We add package dependencies with
usethis::use_package()