Introduction and Principles of reproducibility
Introduction
Programme Overview
What this training is and is not
Chosen topics are: foundational, widely applicable, and transferable conceptually.
It is
- ✅ An introduction to R for those without previous experience
- ✅ About using RStudio Projects and good practice for code and project documentation and organisation
- ✅ An introduction to the tidyverse
It is not
- ❌ An introduction to statistics
- ❌ Magic
Learning Objectives
After this workshop the successful learner will be able to:
- Explain the rationale for scripting analysis
- Find their way around the RStudio windows
- Create and plot data using ggplot
- Know how to load packages
- Understand what is meant by the working directory, absolute and relative paths and be able to apply these concepts to data import
- Summarise data in a single group or in multiple groups
- Develop highly organised analyses including well-commented scripts that can be understood by future you and others
Principles of reproducibility
What is reproducibility?
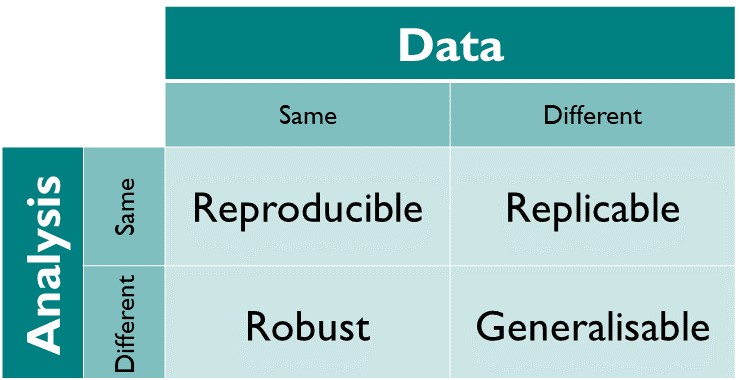
Reproducible: Same data + same analysis = identical results
“…obtaining consistent results using the same input data; computational steps, methods, and code; and conditions of analysis. This definition is synonymous with”computational reproducibility” (National Academies of Sciences et al. 2019). This is what we are concentrating on in the Supporting Information.
Replicable: Different data + same analysis = qualitatively similar results. The work is not dependent on the specificities of the data.
Robust: Same data + different analysis = qualitatively similar or identical results. The work is not dependent on the specificities of the analysis.
Generalisable: Different data + different analysis = qualitatively similar results and same conclusions.
Why does reproducibility matter?
Many high profile cases of work which did not reproduce e.g. Anil Potti unravelled by Baggerly and Coombes (2009)
Five selfish reasons to work reproducibly (Markowetz 2015). Alternatively, see the very entertaining talk
Will become standard in Science and publishing e.g OECD Global Science Forum Building digital workforce capacity and skills for data-intensive science (OECD Global Science Forum 2020)
How to achieve reproducibility
Reproducibility is a continuum. Some is better than none!
Script everything
Organisation: Project-oriented workflows with file and folder structure, naming things
Code: follow a consistent style, organise into sections and scripts (be modular), Code algorithmically
Documentation: Readme files, code comments, metadata,
More advanced: version, control, continuous integration and testing
Rationale for scripting
Science is the generation of ideas, designing work to test them and reporting the results.
We ensure laboratory and field work is replicable, robust and generalisable by planning and recording in lab books and using standard protocols. Repeating results is still hard.
Workflows for computational projects, and the data analysis and reporting of other work can, and should, be 100% reproducible!
Scripting is the way to achieve this.
Project-oriented workflow
use folders to organise your work
you are aiming for structured, systematic and repeatable.
inputs and outputs should be clearly identifiable from structure and/or naming
Further reading
- Data Organization in Spreadsheets (Broman and Woo 2018)
- Ten simple rules for reproducible computational research (Sandve et al. 2013)
- Best practices for scientific computing (Wilson et al. 2014)
- Good enough practices in scientific computing (Wilson et al. 2017)
- Excuse Me, Do You Have a Moment to Talk About Version Control? (Bryan 2018)
Pages made with R (R Core Team 2025), Quarto (Allaire et al. 2024), knitr (Xie 2024, 2015, 2014), kableExtra (Zhu 2024)