usethis::use_course(url = "3mmaRand/structure-analysis", destdir = ".")Structure Data Analysis for Group Project
Extra help to make links with Python, R and Quarto
Programmatic protein structure analysis
There is an RStudio project containing a Quarto version of the the Antibody Mimetics Workshop by Michael Plevin & Jon Agirre. Instructions to obtain the RStudio project are at the bottom of this document after the set up instructions.
You might find RStudio useful for Python because you are already familiar with it. It is also a good way to create Quarto documents with code chunks in more than one language. Quarto documents can be used in RStudio, VS Code or Jupyter notebooks
Some set up is required before you will be able to execute code in antibody_mimetics_workshop_3.qmd. This in contrast to the Colab notebook which is a cloud-based Jupyter notebook and does not require any set up (except installing packages).
Set up
🎬 If using your own machine, install Python from https://www.python.org/downloads/. This should not be necessary if you are using a university machine where Python is already installed.
🎬 If using your own machine and you did not install Quarto in the Core 1 workshop, install it now from https://quarto.org/docs/get-started/. This should not be necessary if you are using a university machine where quarto is already installed.
🎬 Open RStudio and check you are using a “Git bash” Terminal: Tools | Global Options| Terminal | New Terminal opens with… . If the option to choose Git bash, you will need to install Git from https://git-scm.com/downloads. Quit RStudio first. This should not be necessary if you are using a university machine where Git bash is already installed.
🎬 If on your own machine: In RStudio, install the quarto and the recticulate packages. This should not be necessary if you are using a university machine where these packages are already installed.
🎬 Whether you are using your own machine or a university machine, you need to install some python packages. In RStudio and go to the Terminal window (behind the Console window). Run the following commands in the Terminal window:
python -m pip install --upgrade pip setuptools wheelYou may get these warnings about scripts not being on the path. You can ignore these.
WARNING: The script wheel.exe is installed in 'C:\Users\er13\AppData\Roaming\Python\Python39\Scripts' which is not on PATH.
Consider adding this directory to PATH or, if you prefer to suppress this warning, use --no-warn-script-location.
WARNING: The scripts pip.exe, pip3.11.exe, pip3.9.exe and pip3.exe are installed in 'C:\Users\er13\AppData\Roaming\Python\Python39\Scripts' which is not on PATH.
Consider adding this directory to PATH or, if you prefer to suppress this warning, use --no-warn-script-location.
ERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.
spyder 5.1.5 requires pyqt5<5.13, which is not installed.
spyder 5.1.5 requires pyqtwebengine<5.13, which is not installed.
conda-repo-cli 1.0.4 requires pathlib, which is not installed.
anaconda-project 0.10.2 requires ruamel-yaml, which is not installed.
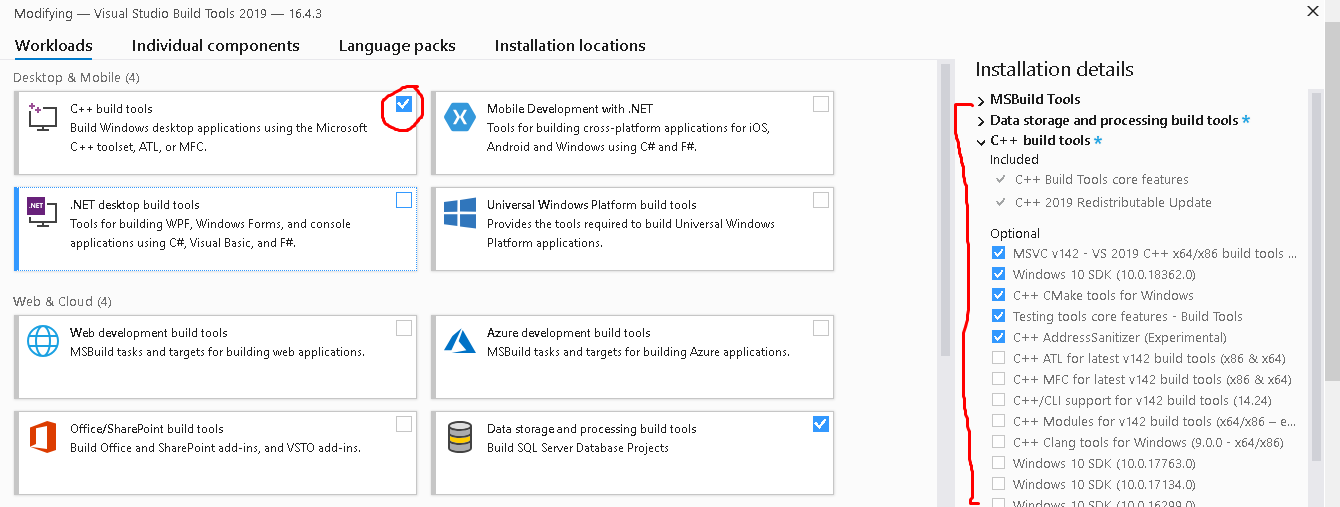
Successfully installed pip-23.3.1 setuptools-69.0.2 wheel-0.41.3python -m pip install session_infopython -m pip install wgetpython -m pip install gemmiNote: On my windows laptop at home, I also had to install C++ Build Tools to be able to install the gemmi python package. If this is true for you, you will get a fail message telling you to install C++ build tools if you need them. These are from https://visualstudio.microsoft.com/visual-cpp-build-tools/ You need to check the Workloads tab and select C++ build tools.
You can then install the gemmi package again.
I think that’s it! You can now download the RStudio project and run each chunk in the quarto document.
There is an example RStudio project here: structure-analysis. You can also download the project as a zip file from there but there is some code that will do that automatically for you. Since this is an RStudio Project, do not run the code from inside a project. You may want to navigate to a particular directory or edit the destdir:
You can agree to deleting the zip. You should find RStudio restarts and you have a new project called structure-analysis-xxxxxx. The xxxxxx is a commit reference - you do not need to worry about that, it is just a way to tell you which version of the repo you downloaded.
You should be able to open the antibody_mimetics_workshop_3.qmd file and run each chunk. You can also knit the document to html.