culture <- read_csv("data-raw/culture.csv")14 One-way ANOVA and Kruskal-Wallis
You are reading a work in progress. This page is compete but needs final proof reading.
14.1 Overview
In the last chapter, we learnt how to use and interpret the general linear model when the x variable was categorical with two groups. You will now extend that to situations when there are more than two groups. This is often known as the one-way ANOVA (analysis of variance). We will also learn about the Kruskal-Wallis test (Kruskal and Wallis 1952), a non-parametric test that can be used when the assumptions of the general linear model are not met.
We use lm() to carry out a one-way ANOVA. General linear models applied with lm() are based on the normal distribution and known as parametric tests because they use the parameters of the normal distribution (the mean and standard deviation) to determine if an effect is significant. Null hypotheses are about a mean or difference between means. The assumptions need to be met for the p-values generated to be accurate.
If the assumptions are not met, we can use the non-parametric equivalent known as the Kruskal-Wallis test. Like other non-parametric tests, the Kruskal-Wallis test :
- is based on the ranks of values rather than the actual values themselves
- has a null hypothesis about the mean rank rather than the mean
- has fewer assumptions and can be used in more situations
- tends to be less powerful than a parametric test when the assumptions are met
The process of using lm() to conduct a one-way ANOVA is very similar to using lm() for a two-sample t-test, but with an important distinction. When we obtain a significant effect of our explanatory variable, it only indicates that at least two group means differ—it does not specify which ones. To determine where the differences lie, we need a post-hoc test.
A post-hoc (“after this”) test is performed after a significant ANOVA result. There are several options for post-hoc tests, and we will use Tukey’s HSD (honestly significant difference) test (Tukey 1949), implemented in the emmeans (Lenth 2023) package.
Post-hoc tests adjust p-values to account for multiple comparisons. A Type I error occurs when we incorrectly reject a true null hypothesis, with a probability of 0.05. Conducting multiple comparisons increases the likelihood of obtaining a significant result by chance. The post-hoc test corrects for this increased risk, ensuring more reliable conclusions.
14.1.1 Model assumptions
The assumptions for a general linear model where the explanatory variable has two or more groups, are the same as for two groups: the residuals are normally distributed and have homogeneity of variance.
If we have a continuous response and a categorical explanatory variable with three or more groups, we usually apply the general linear model with lm() and then check the assumptions, however, we can sometimes tell when a non-parametric test would be more appropriate before that:
- Use common sense - the response should be continuous (or nearly continuous, see Ideas about data: Theory and practice). Consider whether you would expect the response to be continuous
- There should decimal places and few repeated values.
To examine the assumptions after fitting the linear model, we plot the residuals and test them against the normal distribution in the same way as we did for single linear regression.
14.1.2 Reporting
In reporting the result of one-way ANOVA or Kruskal-Wallis test, we include:
-
the significance of the effect
- parametric (GLM): The F-statistic and p-value
- non-parametric (Kruskal-Wallis): The Chi-squared statistic and p-value
-
the direction of effect - which mean/median is greater in a the pairwise commparison
- Post-hoc test
-
the magnitude of effect - how big is the difference between the means/medians
- parametric: the means and standard errors for each group
- non-parametric: the medians for each group
Figures should reflect what you have said in the statements. Ideally they should show both the raw data and the statistical model:
- parametric: means and standard errors
- non-parametric: boxplots with medians and interquartile range
We will explore all of these ideas with some examples.
14.2 🎬 Your turn!
If you want to code along you will need to start a new RStudio project, add a data-raw folder and open a new script. You will also need to load the tidyverse package (Wickham et al. 2019).
14.3 One-way ANOVA
Researchers wanted the determine the best growth medium for growing bacterial cultures. They grew bacterial cultures on three different media formulations and measured the diameter of the colonies. The three formulations were:
- Control - a generic medium as formulated by the manufacturer
- sugar added - the generic medium with added sugar
- sugar and amino acids added - the generic medium with added sugar and amino acids
The data are in culture.csv.
In this scenario, our null hypothesis, \(H_0\), is that there is no difference in colony diameter between the three groups or that group membership has no effect on diameter. This is written as: \(H_0: \beta_1 = \beta_2 = 0\)
This means that none of the group means differ significantly from each other.
Another way of describing, \(H_0\), is to say the variation caused by media is no greater than the the random background variation. This is expressed with the F-statistic, a variance ratio that compares the variance explained by the model to the variance within groups. If \(H_0\) is true, the variance between groups is no greater than the variance within groups. That is, F, \(\frac{Var(between)}{Var(within)}\) equals 1. This is written as: \(H_0: F = 1\)
Both versions of the null hypothesis describe the same idea: that here is no real difference between the groups.
Using \(\beta\) coefficients: This version focuses on the linear model. It says that the group variable has no effect, meaning all groups have the same average value. If the coefficients (\(\beta_1\), \(\beta_2\), etc.) are zero, then the model isn’t capturing any meaningful differences between groups.
Using the F-statistic: This version looks at variance. It compares the variation between groups to the variation within groups. If the F-value is close to 1, it means the differences between groups are about the same as random variation within groups, so there’s no real effect.
Both approaches test the same hypothesis—if we reject the null, we conclude that at least one group is different from the others.
14.3.1 Import and explore
Import the data:
| diameter | medium |
|---|---|
| 11.22 | control |
| 9.35 | control |
| 9.15 | control |
| 10.35 | control |
| 9.63 | control |
| 10.96 | control |
| 10.07 | control |
| 10.40 | control |
| 10.33 | control |
| 9.24 | control |
| 8.90 | sugar added |
| 10.75 | sugar added |
| 11.95 | sugar added |
| 9.85 | sugar added |
| 10.12 | sugar added |
| 10.05 | sugar added |
| 9.60 | sugar added |
| 10.10 | sugar added |
| 10.20 | sugar added |
| 10.88 | sugar added |
| 10.45 | sugar and amino acids added |
| 13.19 | sugar and amino acids added |
| 11.84 | sugar and amino acids added |
| 13.35 | sugar and amino acids added |
| 11.22 | sugar and amino acids added |
| 9.86 | sugar and amino acids added |
| 10.27 | sugar and amino acids added |
| 10.62 | sugar and amino acids added |
| 11.78 | sugar and amino acids added |
| 11.43 | sugar and amino acids added |
The Response variable is colony diameters in millimetres and we would expect it to be continuous. The Explanatory variable is type of media and is categorical with 3 groups. It is known “one-way ANOVA” or “one-factor ANOVA” because there is only one explanatory variable. It would still be one-way ANOVA if we had 4, 20 or 100 media.
These data are in tidy format (Wickham 2014) - all the diameter values are in one column with another column indicating the media. This means they are well formatted for analysis and plotting.
In the first instance it is sensible to create a rough plot of our data. This is to give us an overview and help identify if there are any issues like missing or extreme values. It also gives us idea what we are expecting from the analysis which will make it easier for us to identify if we make some mistake in applying that analysis.
Violin plots (geom_violin(), see Figure 14.1), box plots (geom_boxplot()) or scatter plots (geom_point()) all make good choices for exploratory plotting and it does not matter which of these you choose.
ggplot(data = culture,
aes(x = medium, y = diameter)) +
geom_violin()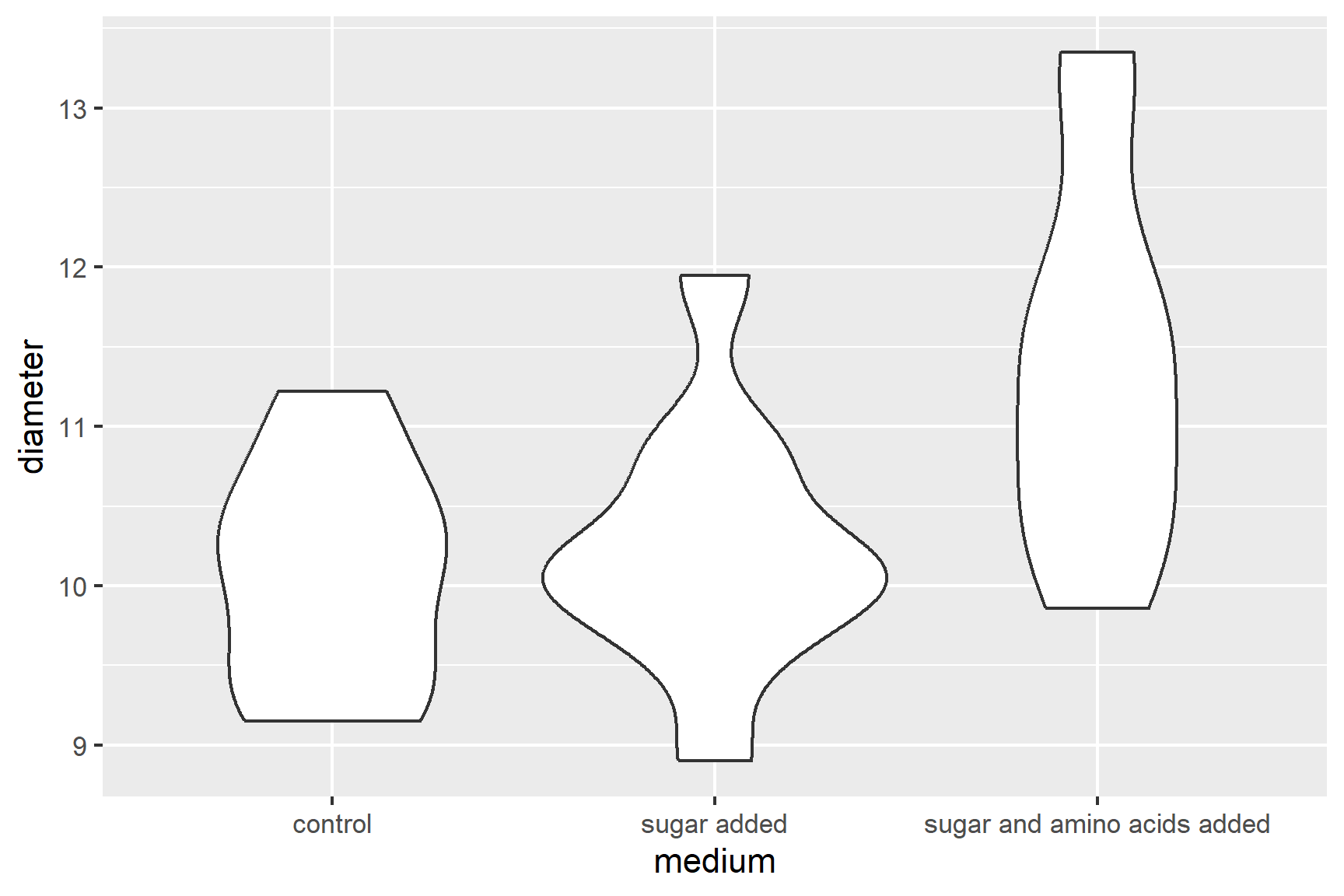
R will order the groups alphabetically by default.
The figure suggests that adding sugar and amino acids to the medium increases the diameter of the colonies.
Summarising the data for each medium is the next sensible step. The most useful summary statistics are the means, standard deviations, sample sizes and standard errors. I recommend the group_by() and summarise() approach:
We have save the results to culture_summary so that we can use the means and standard errors in our plot later.
culture_summary
## # A tibble: 3 × 5
## medium mean std n se
## <chr> <dbl> <dbl> <int> <dbl>
## 1 control 10.1 0.716 10 0.226
## 2 sugar added 10.2 0.818 10 0.259
## 3 sugar and amino acids added 11.4 1.18 10 0.373
14.3.2 Apply lm()
We can create a one-way ANOVA model like this:
mod <- lm(data = culture, diameter ~ medium)And examine the model with:
summary(mod)
##
## Call:
## lm(formula = diameter ~ medium, data = culture)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.541 -0.700 -0.080 0.424 1.949
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 10.0700 0.2930 34.370 < 2e-16 ***
## mediumsugar added 0.1700 0.4143 0.410 0.68483
## mediumsugar and amino acids added 1.3310 0.4143 3.212 0.00339 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.9265 on 27 degrees of freedom
## Multiple R-squared: 0.3117, Adjusted R-squared: 0.2607
## F-statistic: 6.113 on 2 and 27 DF, p-value: 0.00646The Estimates in the Coefficients table give:
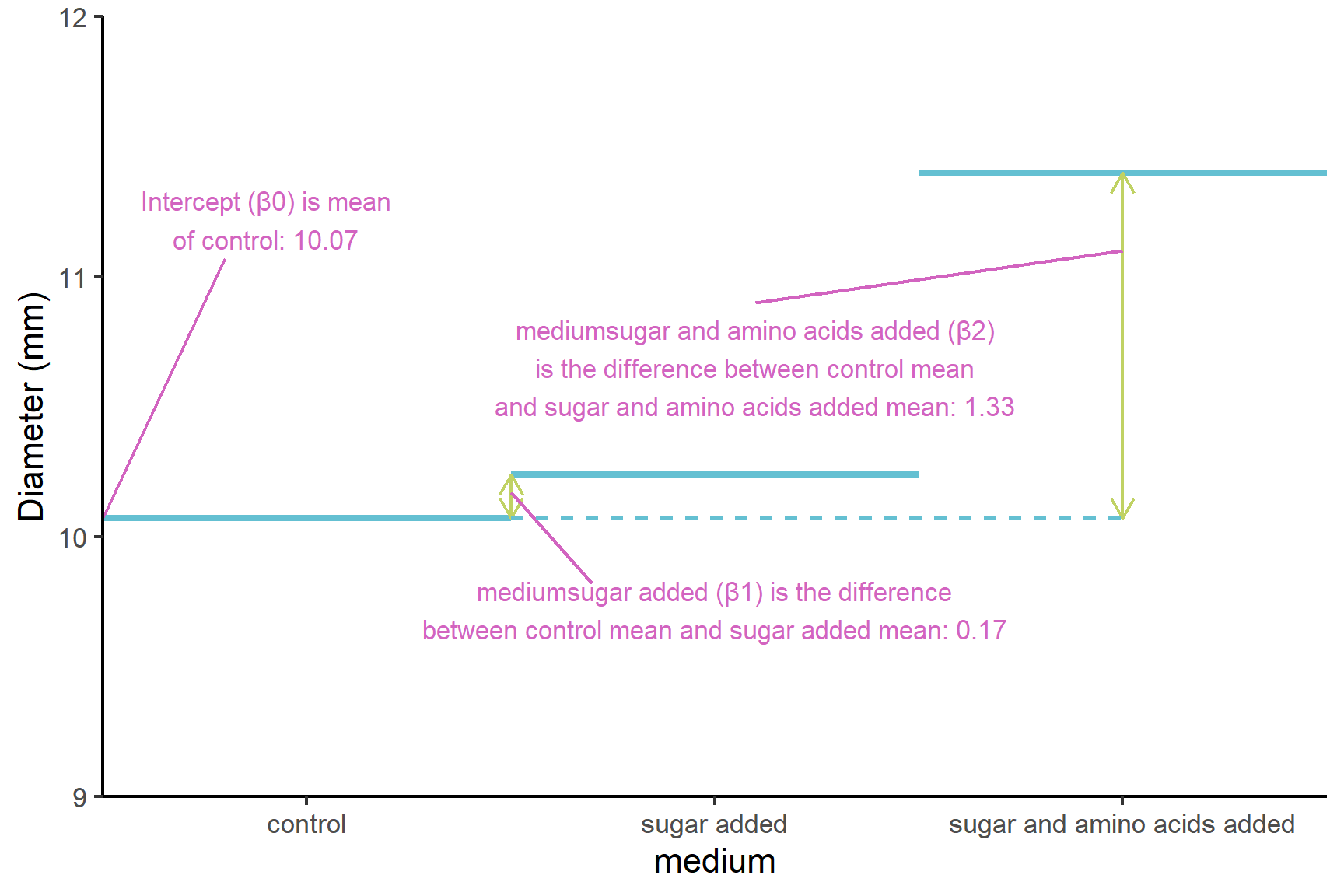
(Intercept)known as \(\beta_0\). The mean of the control group (Figure 14.2). Just as the intercept is the value of the y (the response) when the value of x (the explanatory) is zero in a simple linear regression, this is the value ofdiameterwhen themediumis at its first level. The order of the levels is alphabetical by default.mediumsugar addedknown as \(\beta_1\). This is what needs to be added to the mean of the control group to get the mean of the ‘medium sugar added’ group (Figure 13.3). Just as the slope is amount of y that needs to be added for each unit of x in a simple linear regression, this is the amount ofdiameterthat needs to be added when themediumgoes from its first level to its second level (i.e., one unit). Themediumsugar addedestimate is positive so the the ‘medium sugar added’ group mean is higher than the control group meanmediumsugar and amino acids addedknown as \(\beta_2\) is what needs to be added to the mean of the control group to get the mean of the ‘medium sugar and amino acids added’ group (Figure 13.3). Note that it is the amount added to the intercept (the control in this case). Themediumsugar and amino acids addedestimate is positive so the the ‘medium sugar and amino acids added’ group mean is higher than the control group mean
If we had more groups, we would have more estimates and all would be compared to the control group mean.
The p-values on each line are tests of whether that coefficient is different from zero.
-
(Intercept) 10.0700 0.2930 34.370 < 2e-16 ***tells us that the control group mean is significantly different from zero. This is not a very interesting, it just means the control colonies have a diameter. -
mediumsugar added 0.1700 0.4143 0.410 0.68483tells us that the ‘medium sugar added’ group mean is not significantly different from the control group mean. -
mediumsugar and amino acids added 1.3310 0.4143 3.212 0.00339 **tells us that the ‘medium sugar and amino acids added’ group mean is significantly different from the control group mean.
Note: none of this output tells us whether the medium sugar and amino acids added’ group mean is significantly different from the ‘medium sugar added’ group mean. We need to do a post-hoc test for that.
The F value and p-value in the last line are a test of whether the model as a whole explains a significant amount of variation in the response variable.
The ANOVA is significant but this only tells us that growth medium matters, meaning at least two of the means differ. To find out which means differ, we need a post-hoc test. A post-hoc (“after this”) test is done after a significant ANOVA test. There are several possible post-hoc tests and we will be using Tukey’s HSD (honestly significant difference) test (Tukey 1949) implemented in the emmeans (Lenth 2023) package.
We need to load the package:
Then carry out the post-hoc test:
emmeans(mod, ~ medium) |> pairs()
## contrast estimate SE df t.ratio p.value
## control - sugar added -0.17 0.414 27 -0.410 0.9117
## control - sugar and amino acids added -1.33 0.414 27 -3.212 0.0092
## sugar added - sugar and amino acids added -1.16 0.414 27 -2.802 0.0244
##
## P value adjustment: tukey method for comparing a family of 3 estimatesEach row is a comparison between the two means in the ‘contrast’ column. The ‘estimate’ column is the difference between those means and the ‘p.value’ indicates whether that difference is significant.
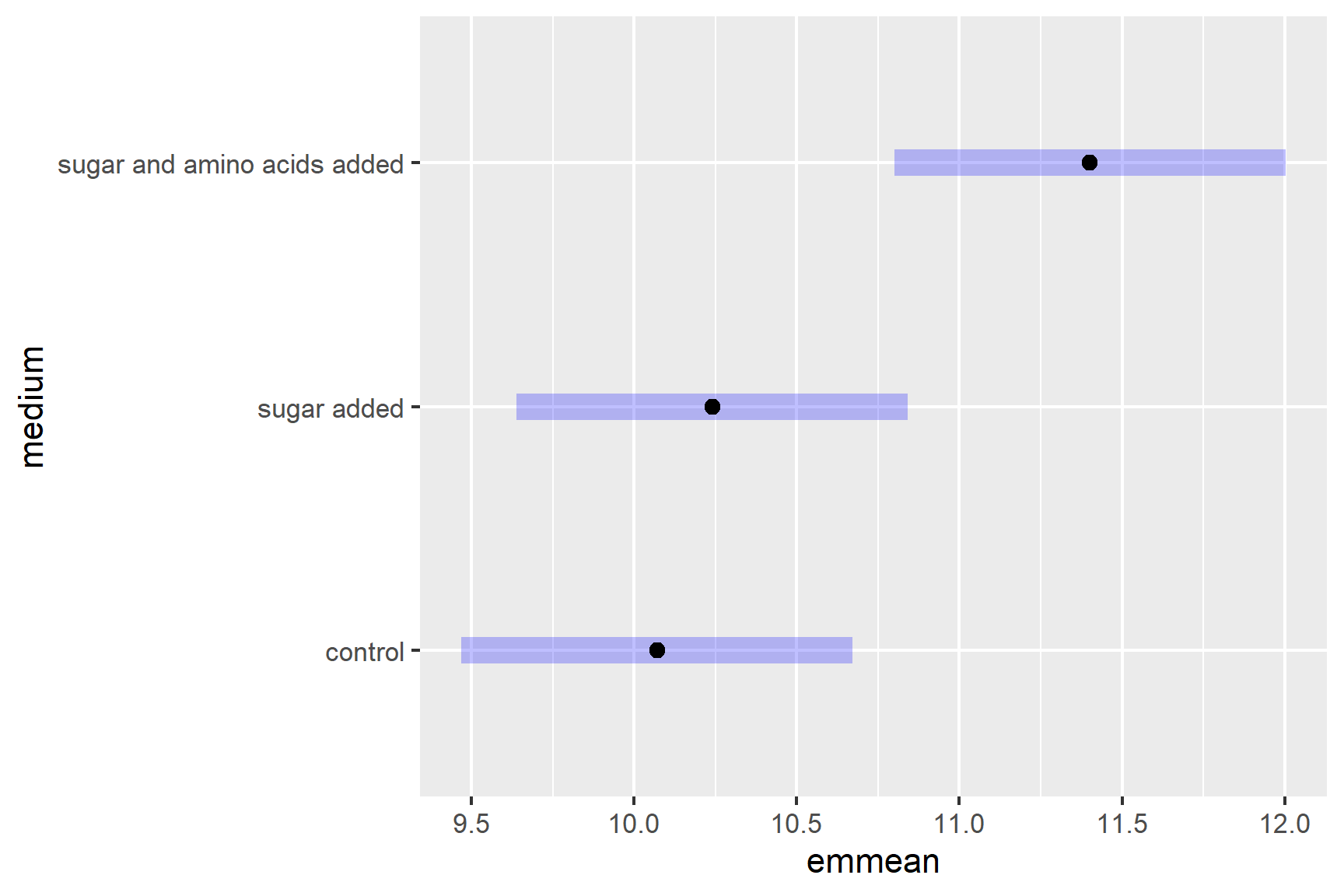
A plot can be used to visualise the result of the post-hoc which can be especially useful when there are very many comparisons.
Where the purple bars overlap, there is no significant difference.
We have found that colony diameters are significantly greater when sugar and amino acids are added but that adding sugar alone does not significantly increase colony diameter.
14.3.3 Check assumptions
Check the assumptions: All general linear models assume the “residuals” are normally distributed and have “homogeneity” of variance.
Our first check of these assumptions is to use common sense: diameter is a continuous and we would expect it to be normally distributed thus we would expect the residuals to be normally distributed thus we would expect the residuals to be normally distributed
We then proceed by plotting residuals. The plot() function can be used to plot the residuals against the fitted values (See Figure 14.3). This is a good way to check for homogeneity of variance.
plot(mod, which = 1)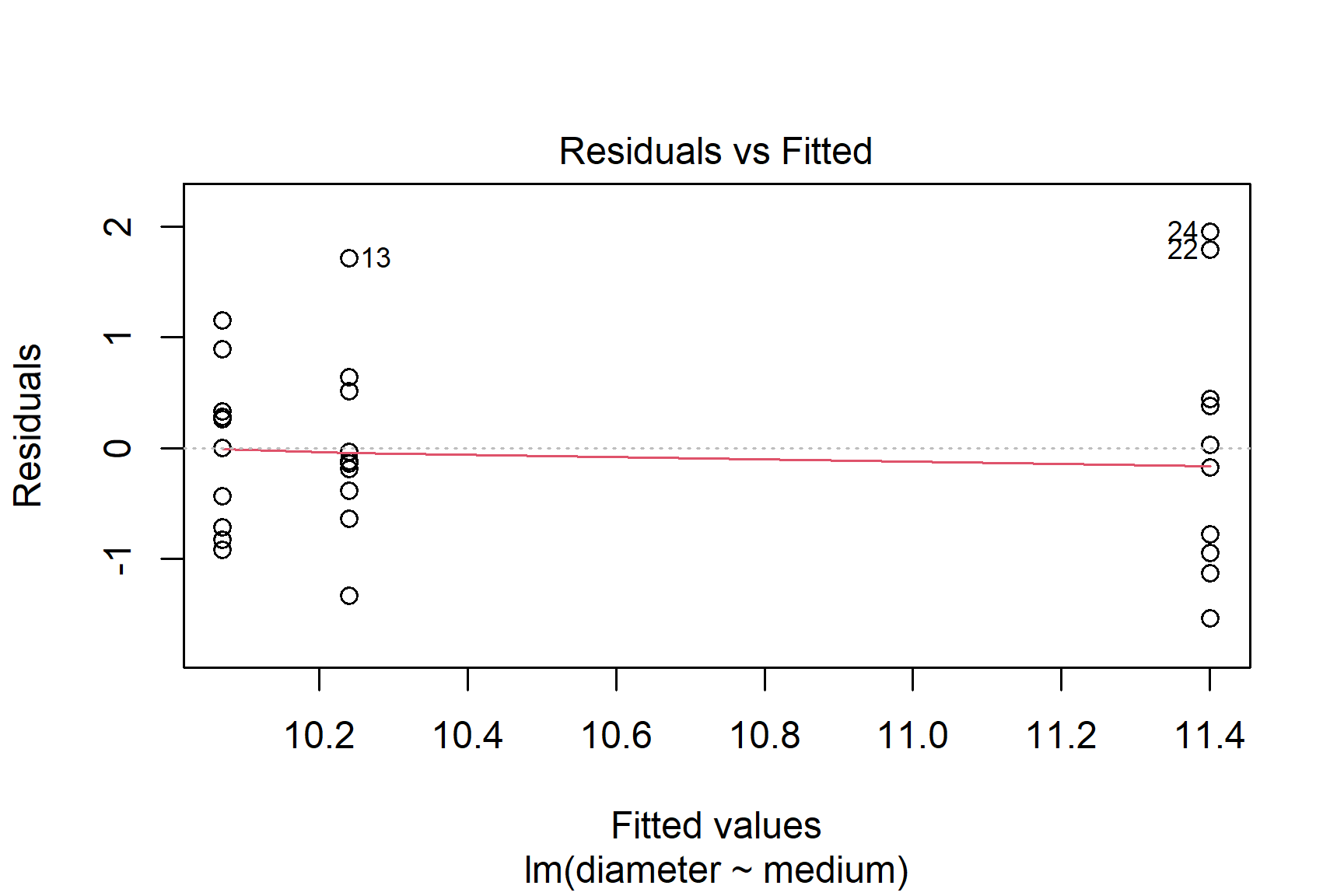
Perhaps the variance is higher for the highest mean?
We can also use a histogram to check for normality (See Figure 14.4).
ggplot(mapping = aes(x = mod$residuals)) +
geom_histogram(bins = 8)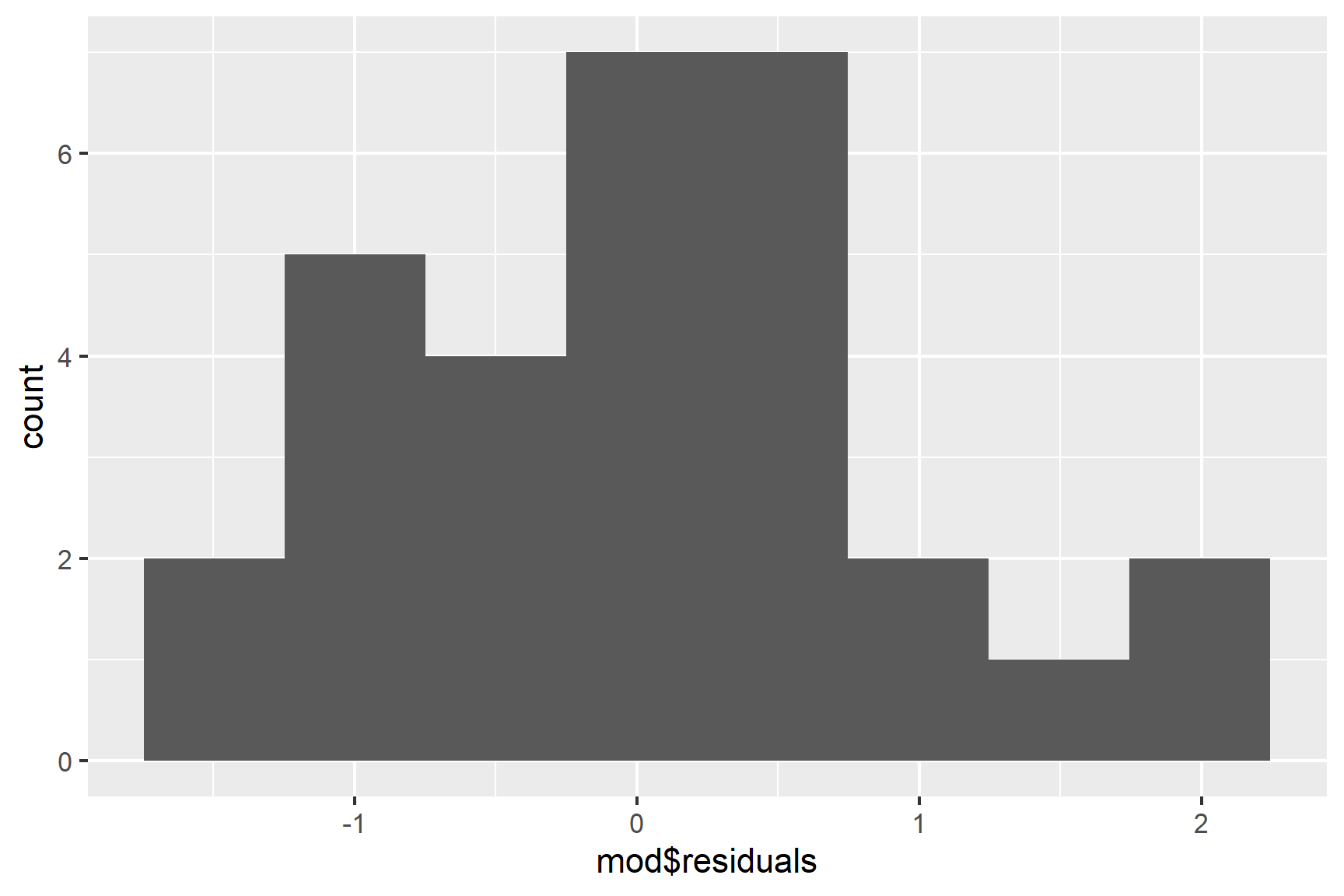
Finally, we can use the Shapiro-Wilk test to test for normality.
shapiro.test(mod$residuals)
##
## Shapiro-Wilk normality test
##
## data: mod$residuals
## W = 0.96423, p-value = 0.3953The p-value is greater than 0.05 so this test of the normality assumption is not significant.
Taken together, these results suggest that the assumptions of normality and homogeneity of variance are probably not violated.
14.3.4 Report
There was a significant effect of media on the diameter of bacterial colonies (F = 6.11; d.f. = 2, 27; p = 0.006). Post-hoc testing with Tukey’s Honestly Significant Difference test (Tukey 1949) revealed the colony diameters were significantly larger when grown with both sugar and amino acids (\(\bar{x} \pm s.e\): 11.4 \(\pm\) 0.37 mm) than with neither (10.2 \(\pm\) 0.26 mm; p = 0.0092) or just sugar (10.1 \(\pm\) 0.23 mm; p = 0.0244). See Figure 14.5.
Code
ggplot() +
geom_point(data = culture, aes(x = medium, y = diameter),
position = position_jitter(width = 0.1, height = 0),
colour = "gray50") +
geom_errorbar(data = culture_summary,
aes(x = medium, ymin = mean - se, ymax = mean + se),
width = 0.3) +
geom_errorbar(data = culture_summary,
aes(x = medium, ymin = mean, ymax = mean),
width = 0.2) +
scale_y_continuous(name = "Diameter (mm)",
limits = c(0, 16.5),
expand = c(0, 0)) +
scale_x_discrete(name = "Medium",
labels = c("Control",
"Sugar added",
"Sugar and amino acids added")) +
annotate("segment", x = 2, xend = 3,
y = 14, yend = 14,
colour = "black") +
annotate("text", x = 2.5, y = 14.5,
label = expression(italic(p)~"= 0.0244")) +
annotate("segment", x = 1, xend = 3,
y = 15.5, yend = 15.5,
colour = "black") +
annotate("text", x = 2, y = 16,
label = expression(italic(p)~"= 0.0092")) +
theme_classic()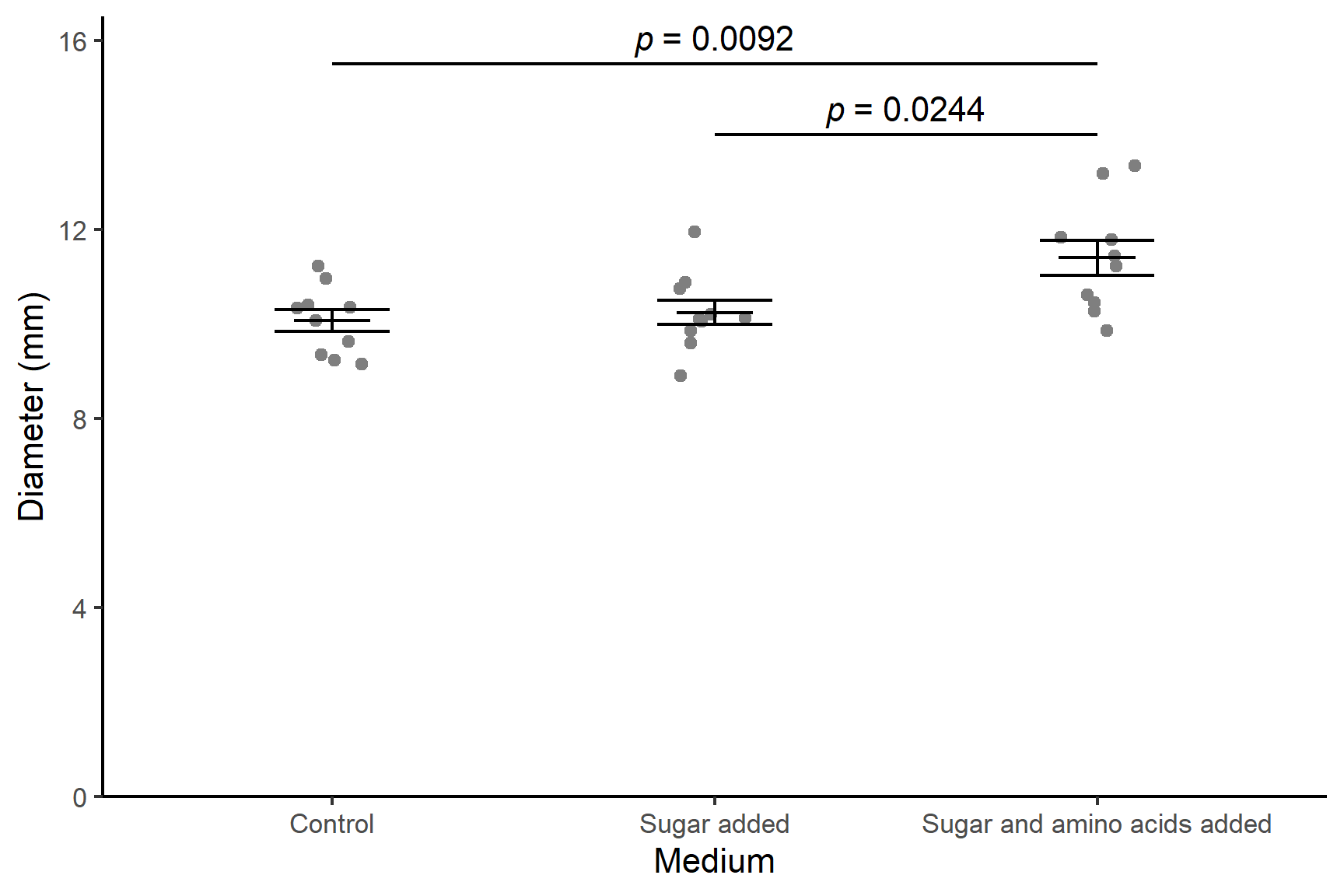
15 Kruskal-Wallis
Our examination of the assumptions revealed a possible violation of the assumption of homogeneity of variance. We might reasonably apply a non-parametric test to this data instead.
The Kruskal-Wallis (Kruskal and Wallis 1952) is non-parametric equivalent of a one-way ANOVA. The general question you have about your data - do these groups differ (or does the medium effect diameter) - is the same, but one of more of the following is true:
- the response variable is not continuous
- the residuals are not normally distributed
- the sample size is too small to tell if they are normally distributed.
- the variance is not homogeneous
Summarising the data using the median and interquartile range is more aligned to the type of analysis than using means and standard deviations:
View the results:
culture_summary
## # A tibble: 3 × 4
## medium median interquartile n
## <chr> <dbl> <dbl> <int>
## 1 control 10.2 0.968 10
## 2 sugar added 10.1 0.713 10
## 3 sugar and amino acids added 11.3 1.33 10
15.0.1 Apply kruskal.test()
We pass the dataframe and variables to kruskal.test() in the same way as we did for lm(). We give the data argument and a “formula” which says diameter ~ medium meaning “explain diameter by medium”.
kruskal.test(data = culture, diameter ~ medium)
##
## Kruskal-Wallis rank sum test
##
## data: diameter by medium
## Kruskal-Wallis chi-squared = 8.1005, df = 2, p-value = 0.01742The result of the test is given on this line: Kruskal-Wallis chi-squared = 8.1005, df = 2, p-value = 0.01742. Chi-squared is the test statistic. The p-value is less than 0.05 meaning there is a significant effect of medium on diameter.
Notice that the p-value is a little larger than for the ANOVA. This is because non-parametric tests are generally more conservative (less powerful) than their parametric equivalents.
A significant Kruskal-Wallis tells us at least two of the groups differ but where do the differences lie? The Dunn test (Dunn 1964) is a post-hoc multiple comparison test for a significant Kruskal-Wallis. It is available in the package FSA (Ogle et al. 2023)
Load the package using:
Then run the post-hoc test with:
dunnTest(data = culture, diameter ~ medium)
## Comparison Z P.unadj P.adj
## 1 control - sugar added -0.2159242 0.82904681 0.82904681
## 2 control - sugar and amino acids added -2.5656880 0.01029714 0.03089142
## 3 sugar added - sugar and amino acids added -2.3497637 0.01878533 0.03757066The P.adj column gives p-value for the comparison listed in the first column. Z is the test statistic. The p-values are a little larger for the control - sugar and amino acids added comparison and the sugar added - sugar and amino acids added comparison but they are still less than 0.05. This means our conclusions are the same as for the ANOVA.
15.0.2 Report
There is a significant effect of media on the diameter of bacterial colonies (Kruskal-Wallis: chi-squared = 6.34; df = 2; p-value = 0.042) with colonies growing significantly better when both sugar and amino acids are added to the medium. Post-hoc testing with the Dunn test (Dunn 1964) revealed the colony diameters were significantly larger when grown with both sugar and amino acids (median = 11.3 mm) than with neither (median = 10.2 mm; p = 0.031) or just sugar (median = 10.2 mm; p = 0.038). See Figure 15.1.
Code
ggplot(data = culture, aes(x = medium, y = diameter)) +
geom_boxplot() +
scale_y_continuous(name = "Diameter (mm)",
limits = c(0, 16.5),
expand = c(0, 0)) +
scale_x_discrete(name = "Medium",
labels = c("Control",
"Sugar added",
"Sugar and amino acids added")) +
annotate("segment", x = 2, xend = 3,
y = 14, yend = 14,
colour = "black") +
annotate("text", x = 2.5, y = 14.5,
label = expression(italic(p)~"= 0.038")) +
annotate("segment", x = 1, xend = 3,
y = 15.5, yend = 15.5,
colour = "black") +
annotate("text", x = 2, y = 16,
label = expression(italic(p)~"= 0.031")) +
theme_classic()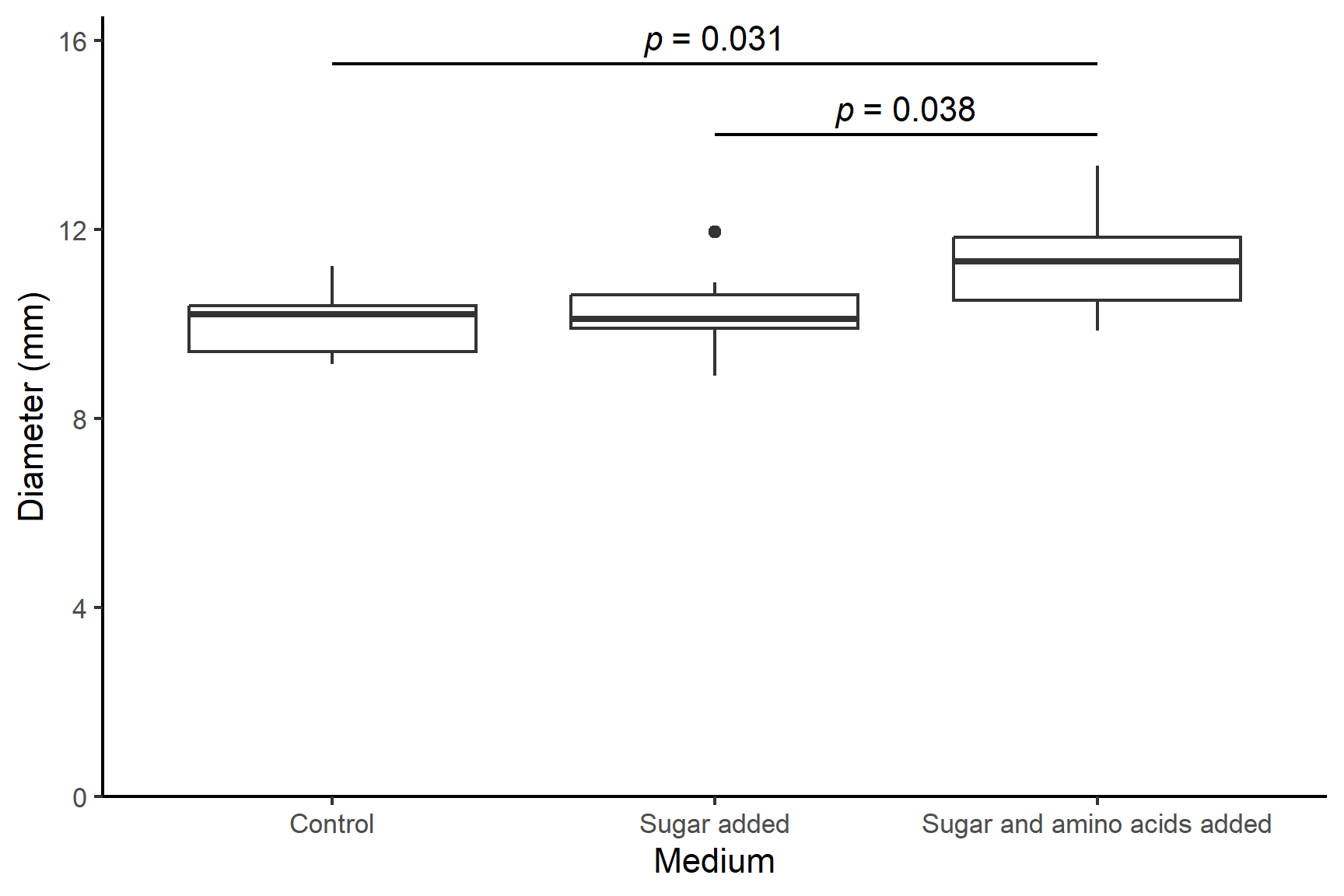
16 Summary
A linear model with one explanatory variable with two or more groups is also known as a one-way ANOVA.
We estimate the coefficients (also called the parameters) of the model. For a one-way ANOVA with three groups these are the mean of the first group, \(\beta_0\), the difference between the means of the first and second groups, \(\beta_1\), and the difference between the means of the first and third groups, \(\beta_2\). We test whether the parameters differ significantly from zero
We can use
lm()to one-way ANOVA in R.In the output of
lm()the coefficients are listed in a table in the Estimates column. The p-value for each coefficient is in the test of whether it differs from zero. At the bottom of the output there is an \(F\) test of the model overall. Now we have more than two parameters, this is different from the test on any one parameter. The R-squared value is the proportion of the variance in the response variable that is explained by the model. It tells us is the explanatory variable is useful in predicting the response variable overall.When the \(F\) test is significant there is a significant effect of the explanatory variable on the response variable. To find out which means differ, we need a post-hoc test. Here we use Tukey’s HSD applied with the
emmeans()andpairs()functions from theemmeanspackage. Post-hoc tests make adjustments to the p-values to account for the fact that we are doing multiple tests.The assumptions of the general linear model are that the residuals are normally distributed and have homogeneity of variance. A residual is the difference between the predicted value and the observed value.
We examine a histogram of the residuals and use the Shapiro-Wilk normality test to check the normality assumption. We check the variance of the residuals is the same for all fitted values with a residuals vs fitted plot.
If the assumptions are not met, we can use the Kruskal-Wallis test applied with
kruskal.test()in R and follow it with The Dunn test applied withdunnTest()in the packageFSA.When reporting the results of a test we give the significance, direction and size of the effect. Our figures and the values we give should reflect the type of test we have used. We use means and standard errors for parametric tests and medians and interquartile ranges for non-parametric tests. We also give the test statistic, the degrees of freedom (parametric) or sample size (non-parametric) and the p-value. We annotate our figures with the p-value, making clear which comparison it applies to.